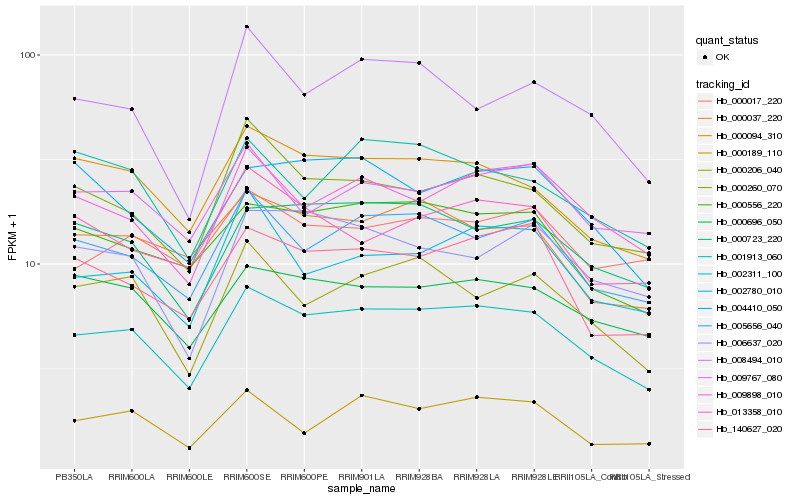
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002311_100 |
0.0 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 11-like [Jatropha curcas] |
| 2 |
Hb_005656_040 |
0.0669826481 |
desease resistance |
Gene Name: T2SE |
ATP-dependent RNA helicase, putative [Ricinus communis] |
| 3 |
Hb_000556_220 |
0.0717289491 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 2-like [Jatropha curcas] |
| 4 |
Hb_000723_220 |
0.0733462723 |
- |
- |
PREDICTED: uncharacterized protein LOC105642234 [Jatropha curcas] |
| 5 |
Hb_000094_310 |
0.0757266 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g75140 [Jatropha curcas] |
| 6 |
Hb_009898_010 |
0.076468289 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105645502 [Jatropha curcas] |
| 7 |
Hb_000696_050 |
0.0784770594 |
- |
- |
PREDICTED: CSC1-like protein At4g35870 [Jatropha curcas] |
| 8 |
Hb_140627_020 |
0.0802906507 |
- |
- |
PREDICTED: uncharacterized protein LOC105637253 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000260_070 |
0.0823498666 |
- |
- |
kinase, putative [Ricinus communis] |
| 10 |
Hb_000206_040 |
0.0845903808 |
- |
- |
PREDICTED: uncharacterized protein LOC105639683 [Jatropha curcas] |
| 11 |
Hb_000037_220 |
0.086537581 |
- |
- |
PREDICTED: vacuolar protein-sorting-associated protein 11 homolog [Jatropha curcas] |
| 12 |
Hb_009767_080 |
0.088788237 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105631983 [Jatropha curcas] |
| 13 |
Hb_002780_010 |
0.0903281323 |
- |
- |
PREDICTED: uncharacterized protein LOC105639237 [Jatropha curcas] |
| 14 |
Hb_008494_010 |
0.0909887264 |
- |
- |
PREDICTED: uncharacterized protein LOC105636015 [Jatropha curcas] |
| 15 |
Hb_006637_020 |
0.0923179416 |
- |
- |
PREDICTED: uncharacterized protein LOC105642234 [Jatropha curcas] |
| 16 |
Hb_000017_220 |
0.0928633221 |
- |
- |
PREDICTED: uncharacterized protein LOC105645768 [Jatropha curcas] |
| 17 |
Hb_001913_060 |
0.0941962626 |
- |
- |
PREDICTED: splicing factor 3B subunit 3-like [Jatropha curcas] |
| 18 |
Hb_004410_050 |
0.094423904 |
- |
- |
PREDICTED: uncharacterized TPR repeat-containing protein At1g05150-like [Jatropha curcas] |
| 19 |
Hb_013358_010 |
0.0944326914 |
- |
- |
hypothetical protein JCGZ_07102 [Jatropha curcas] |
| 20 |
Hb_000189_110 |
0.0946209795 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g04750, mitochondrial [Jatropha curcas] |