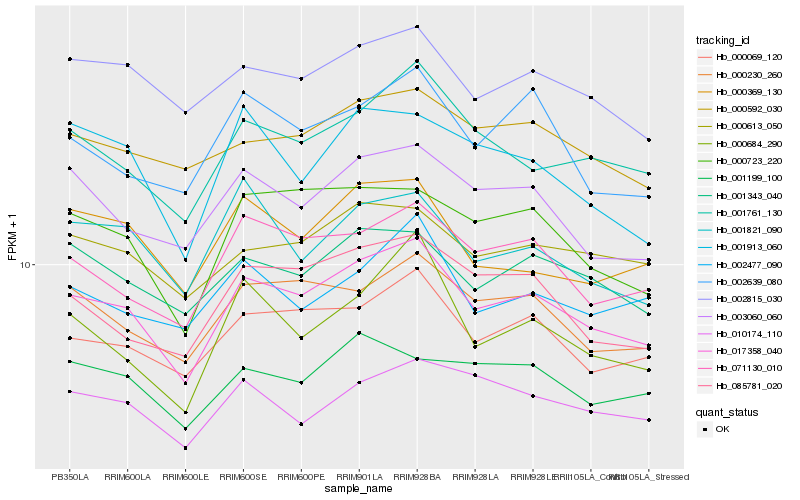
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_017358_040 |
0.0 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 6-like [Jatropha curcas] |
| 2 |
Hb_000069_120 |
0.0607136993 |
- |
- |
PREDICTED: uncharacterized protein LOC105124742 isoform X1 [Populus euphratica] |
| 3 |
Hb_001761_130 |
0.0660455673 |
- |
- |
PREDICTED: uncharacterized protein LOC105646118 isoform X1 [Jatropha curcas] |
| 4 |
Hb_071130_010 |
0.0673154915 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000230_260 |
0.0674315034 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RF298 [Jatropha curcas] |
| 6 |
Hb_000723_220 |
0.06905095 |
- |
- |
PREDICTED: uncharacterized protein LOC105642234 [Jatropha curcas] |
| 7 |
Hb_003060_060 |
0.0720362271 |
- |
- |
PREDICTED: protein transport protein SEC31 homolog B [Jatropha curcas] |
| 8 |
Hb_085781_020 |
0.0722375577 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 1 [Jatropha curcas] |
| 9 |
Hb_001343_040 |
0.0735654504 |
- |
- |
PREDICTED: uncharacterized protein LOC105638555 [Jatropha curcas] |
| 10 |
Hb_002815_030 |
0.0748210452 |
- |
- |
hypothetical protein CISIN_1g0095162mg, partial [Citrus sinensis] |
| 11 |
Hb_000613_050 |
0.0773032755 |
- |
- |
PREDICTED: uncharacterized protein LOC105641544 isoform X2 [Jatropha curcas] |
| 12 |
Hb_002639_080 |
0.0794308619 |
- |
- |
PREDICTED: large proline-rich protein BAG6 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000592_030 |
0.0801119437 |
- |
- |
PREDICTED: uncharacterized protein LOC100853969 isoform X1 [Vitis vinifera] |
| 14 |
Hb_000369_130 |
0.0824349313 |
- |
- |
PREDICTED: H/ACA ribonucleoprotein complex subunit 4 [Jatropha curcas] |
| 15 |
Hb_010174_110 |
0.0826822498 |
- |
- |
hypothetical protein JCGZ_17880 [Jatropha curcas] |
| 16 |
Hb_000684_290 |
0.0851209735 |
- |
- |
ubiquitin protein ligase E3a, putative [Ricinus communis] |
| 17 |
Hb_002477_090 |
0.0852476352 |
- |
- |
PREDICTED: uncharacterized protein LOC105631409 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001199_100 |
0.0855506958 |
- |
- |
PREDICTED: uncharacterized protein LOC105639813 [Jatropha curcas] |
| 19 |
Hb_001821_090 |
0.0861794419 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 2 [Jatropha curcas] |
| 20 |
Hb_001913_060 |
0.0862440075 |
- |
- |
PREDICTED: splicing factor 3B subunit 3-like [Jatropha curcas] |