| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
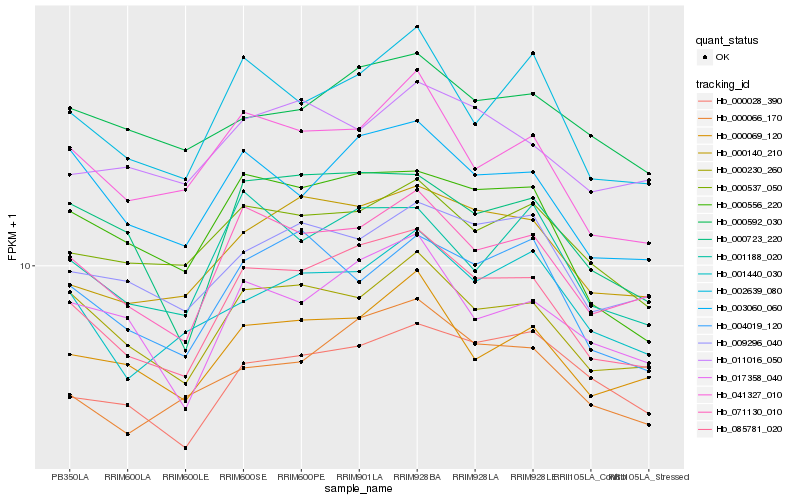
Hb_085781_020 |
0.0 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 1 [Jatropha curcas] |
| 2 |
Hb_071130_010 |
0.0509641212 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000230_260 |
0.0621025923 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RF298 [Jatropha curcas] |
| 4 |
Hb_000140_210 |
0.0638534914 |
- |
- |
sec10, putative [Ricinus communis] |
| 5 |
Hb_000066_170 |
0.0674078355 |
- |
- |
PREDICTED: phosphatidylinositol-3-phosphatase myotubularin-1 isoform X2 [Jatropha curcas] |
| 6 |
Hb_003060_060 |
0.0678609202 |
- |
- |
PREDICTED: protein transport protein SEC31 homolog B [Jatropha curcas] |
| 7 |
Hb_002639_080 |
0.0680533235 |
- |
- |
PREDICTED: large proline-rich protein BAG6 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000556_220 |
0.0681627787 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 2-like [Jatropha curcas] |
| 9 |
Hb_000069_120 |
0.0702786436 |
- |
- |
PREDICTED: uncharacterized protein LOC105124742 isoform X1 [Populus euphratica] |
| 10 |
Hb_041327_010 |
0.0704668755 |
- |
- |
PREDICTED: la-related protein 1A isoform X1 [Jatropha curcas] |
| 11 |
Hb_017358_040 |
0.0722375577 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 6-like [Jatropha curcas] |
| 12 |
Hb_000723_220 |
0.0726784902 |
- |
- |
PREDICTED: uncharacterized protein LOC105642234 [Jatropha curcas] |
| 13 |
Hb_001188_020 |
0.0752479165 |
- |
- |
PREDICTED: E3 SUMO-protein ligase SIZ1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001440_030 |
0.0780557693 |
- |
- |
PREDICTED: exocyst complex component SEC8 [Jatropha curcas] |
| 15 |
Hb_000537_050 |
0.0785515886 |
- |
- |
AP-2 complex subunit beta-1, putative [Ricinus communis] |
| 16 |
Hb_009296_040 |
0.0786728013 |
- |
- |
hypothetical protein L484_007435 [Morus notabilis] |
| 17 |
Hb_004019_120 |
0.0789732953 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 1 [Jatropha curcas] |
| 18 |
Hb_000028_390 |
0.0823799837 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 3-like [Jatropha curcas] |
| 19 |
Hb_000592_030 |
0.0875490106 |
- |
- |
PREDICTED: uncharacterized protein LOC100853969 isoform X1 [Vitis vinifera] |
| 20 |
Hb_011016_050 |
0.0891454144 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI7 isoform X1 [Jatropha curcas] |