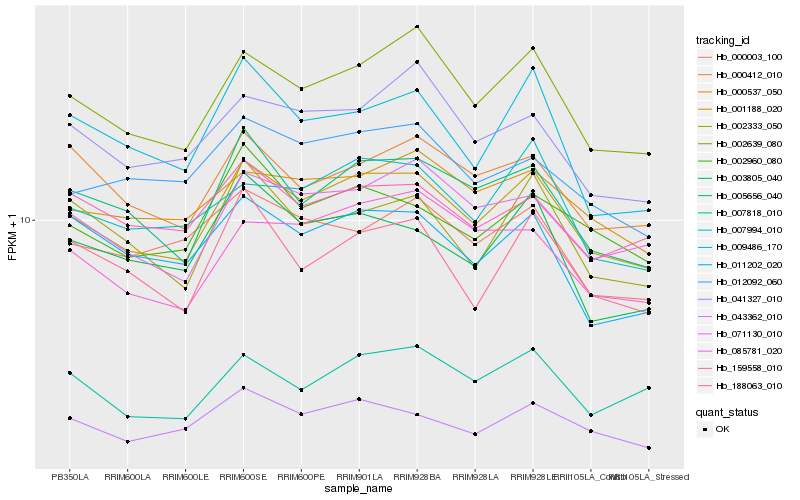
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001188_020 |
0.0 |
- |
- |
PREDICTED: E3 SUMO-protein ligase SIZ1 isoform X1 [Jatropha curcas] |
| 2 |
Hb_002639_080 |
0.0479061481 |
- |
- |
PREDICTED: large proline-rich protein BAG6 isoform X1 [Jatropha curcas] |
| 3 |
Hb_005656_040 |
0.0598506734 |
desease resistance |
Gene Name: T2SE |
ATP-dependent RNA helicase, putative [Ricinus communis] |
| 4 |
Hb_071130_010 |
0.0636342218 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_043362_010 |
0.0651560082 |
- |
- |
hypothetical protein JCGZ_23765 [Jatropha curcas] |
| 6 |
Hb_007994_010 |
0.0686077572 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR-RED ELONGATED HYPOCOTYL 3 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000537_050 |
0.0704620493 |
- |
- |
AP-2 complex subunit beta-1, putative [Ricinus communis] |
| 8 |
Hb_002960_080 |
0.070847499 |
transcription factor |
TF Family: IWS1 |
PREDICTED: uncharacterized protein LOC105635322 [Jatropha curcas] |
| 9 |
Hb_012092_060 |
0.0741613016 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 [Jatropha curcas] |
| 10 |
Hb_000412_010 |
0.0746481863 |
- |
- |
hect ubiquitin-protein ligase, putative [Ricinus communis] |
| 11 |
Hb_085781_020 |
0.0752479165 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 1 [Jatropha curcas] |
| 12 |
Hb_159558_010 |
0.075704395 |
- |
- |
PREDICTED: uncharacterized protein LOC105635846 [Jatropha curcas] |
| 13 |
Hb_041327_010 |
0.0761724983 |
- |
- |
PREDICTED: la-related protein 1A isoform X1 [Jatropha curcas] |
| 14 |
Hb_003805_040 |
0.0764147654 |
- |
- |
Sacsin [Glycine soja] |
| 15 |
Hb_000003_100 |
0.0769634873 |
transcription factor |
TF Family: SNF2 |
PREDICTED: transcription termination factor 2 isoform X5 [Jatropha curcas] |
| 16 |
Hb_002333_050 |
0.0771489808 |
- |
- |
PREDICTED: uncharacterized protein LOC105644854 isoform X1 [Jatropha curcas] |
| 17 |
Hb_009486_170 |
0.0774551698 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 4G [Jatropha curcas] |
| 18 |
Hb_007818_010 |
0.0776639715 |
- |
- |
PREDICTED: transcriptional corepressor SEUSS-like isoform X2 [Gossypium raimondii] |
| 19 |
Hb_188063_010 |
0.077760684 |
- |
- |
hypothetical protein RCOM_1469330 [Ricinus communis] |
| 20 |
Hb_011202_020 |
0.0781480024 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 3 isoform X1 [Jatropha curcas] |