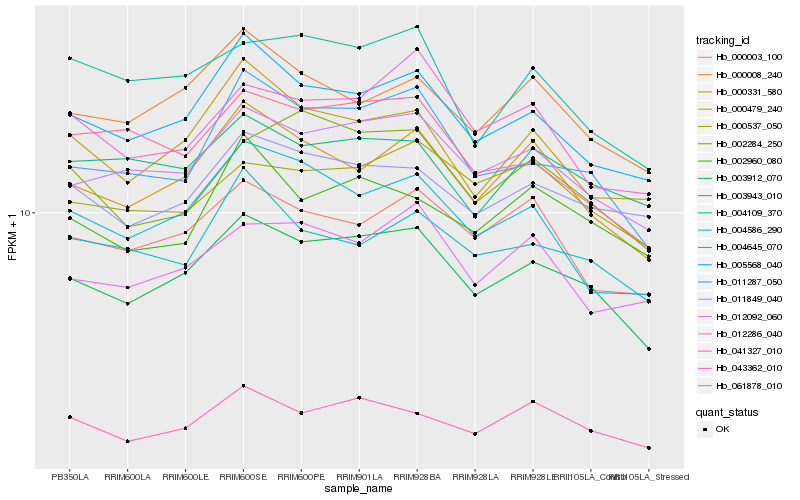
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003912_070 |
0.0 |
- |
- |
PREDICTED: nuclear pore complex protein NUP85 [Jatropha curcas] |
| 2 |
Hb_012092_060 |
0.0492220241 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 [Jatropha curcas] |
| 3 |
Hb_005568_040 |
0.0564260511 |
- |
- |
PREDICTED: uncharacterized protein LOC105628163 isoform X2 [Jatropha curcas] |
| 4 |
Hb_011287_050 |
0.0574799049 |
- |
- |
PREDICTED: uncharacterized protein LOC105644786 [Jatropha curcas] |
| 5 |
Hb_000331_580 |
0.0607407476 |
- |
- |
PREDICTED: programmed cell death protein 4-like [Jatropha curcas] |
| 6 |
Hb_043362_010 |
0.0720006645 |
- |
- |
hypothetical protein JCGZ_23765 [Jatropha curcas] |
| 7 |
Hb_000479_240 |
0.0729191267 |
- |
- |
PREDICTED: probable ubiquitin conjugation factor E4 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000008_240 |
0.0731325766 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_011849_040 |
0.0732614323 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 10 |
Hb_000003_100 |
0.0778038378 |
transcription factor |
TF Family: SNF2 |
PREDICTED: transcription termination factor 2 isoform X5 [Jatropha curcas] |
| 11 |
Hb_004645_070 |
0.0781006641 |
- |
- |
PREDICTED: beta-1,3-galactosyltransferase 15 [Jatropha curcas] |
| 12 |
Hb_002284_250 |
0.0785544438 |
- |
- |
PREDICTED: epidermal growth factor receptor substrate 15-like 1 [Jatropha curcas] |
| 13 |
Hb_012286_040 |
0.0795053926 |
- |
- |
PREDICTED: pre-mRNA-splicing factor RSE1 [Jatropha curcas] |
| 14 |
Hb_000537_050 |
0.0804430525 |
- |
- |
AP-2 complex subunit beta-1, putative [Ricinus communis] |
| 15 |
Hb_002960_080 |
0.0807300627 |
transcription factor |
TF Family: IWS1 |
PREDICTED: uncharacterized protein LOC105635322 [Jatropha curcas] |
| 16 |
Hb_004109_370 |
0.0815562353 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 17 |
Hb_061878_010 |
0.0817493562 |
- |
- |
hypothetical protein CISIN_1g0094072mg, partial [Citrus sinensis] |
| 18 |
Hb_003943_010 |
0.0823697323 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_041327_010 |
0.0826533258 |
- |
- |
PREDICTED: la-related protein 1A isoform X1 [Jatropha curcas] |
| 20 |
Hb_004586_290 |
0.082973692 |
- |
- |
PREDICTED: transport inhibitor response 1-like protein [Jatropha curcas] |