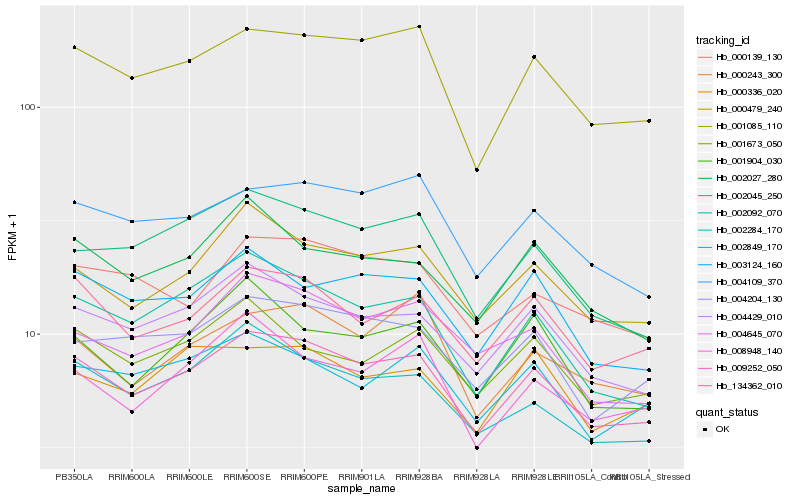
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009252_050 |
0.0 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 2 |
Hb_001085_110 |
0.0485692608 |
- |
- |
T6D22.2 [Arabidopsis thaliana] |
| 3 |
Hb_134362_010 |
0.0532521189 |
- |
- |
PREDICTED: FAS-associated factor 2 [Jatropha curcas] |
| 4 |
Hb_004429_010 |
0.0540776285 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 5 |
Hb_000479_240 |
0.0561173012 |
- |
- |
PREDICTED: probable ubiquitin conjugation factor E4 isoform X2 [Jatropha curcas] |
| 6 |
Hb_004109_370 |
0.0619675259 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 7 |
Hb_004204_130 |
0.0631382996 |
- |
- |
PREDICTED: uncharacterized protein LOC103930912 [Pyrus x bretschneideri] |
| 8 |
Hb_002284_170 |
0.0650766333 |
- |
- |
PREDICTED: serine/threonine-protein kinase/endoribonuclease IRE1b [Jatropha curcas] |
| 9 |
Hb_003124_160 |
0.0654405622 |
- |
- |
dynamin, putative [Ricinus communis] |
| 10 |
Hb_002092_070 |
0.067664529 |
- |
- |
PREDICTED: aluminum-activated malate transporter 9-like [Jatropha curcas] |
| 11 |
Hb_002045_250 |
0.06875339 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP63 isoform X3 [Jatropha curcas] |
| 12 |
Hb_002027_280 |
0.0695474915 |
- |
- |
PREDICTED: sister-chromatid cohesion protein 3 [Jatropha curcas] |
| 13 |
Hb_004645_070 |
0.0705995408 |
- |
- |
PREDICTED: beta-1,3-galactosyltransferase 15 [Jatropha curcas] |
| 14 |
Hb_001673_050 |
0.0711843205 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 8-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_002849_170 |
0.0711885783 |
- |
- |
PREDICTED: uncharacterized protein LOC105642955 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000139_130 |
0.0716225462 |
- |
- |
PREDICTED: chloride channel protein CLC-d isoform X1 [Jatropha curcas] |
| 17 |
Hb_000243_300 |
0.0748532404 |
- |
- |
catalytic, putative [Ricinus communis] |
| 18 |
Hb_001904_030 |
0.0748575977 |
- |
- |
PREDICTED: importin-5 [Jatropha curcas] |
| 19 |
Hb_008948_140 |
0.0758135087 |
- |
- |
PREDICTED: nuclear pore complex protein NUP205 [Jatropha curcas] |
| 20 |
Hb_000336_020 |
0.0758341886 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 2 [Jatropha curcas] |