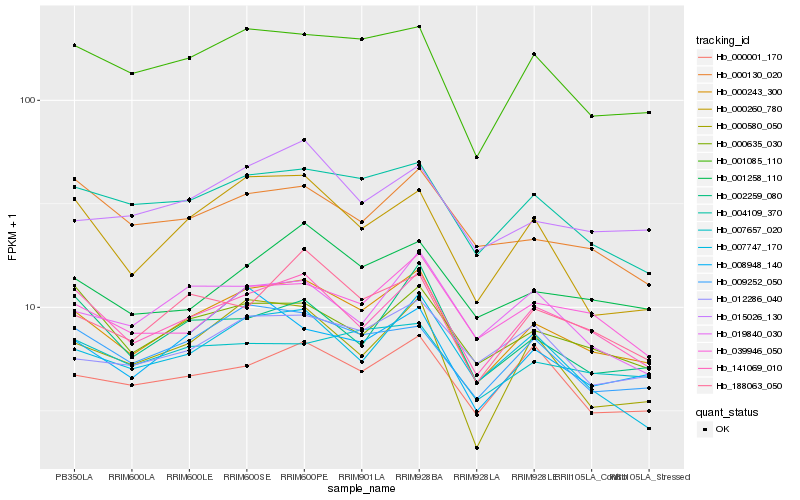
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000243_300 |
0.0 |
- |
- |
catalytic, putative [Ricinus communis] |
| 2 |
Hb_141069_010 |
0.0615399864 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001085_110 |
0.0688661738 |
- |
- |
T6D22.2 [Arabidopsis thaliana] |
| 4 |
Hb_004109_370 |
0.0721908651 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 5 |
Hb_009252_050 |
0.0748532404 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 6 |
Hb_188063_050 |
0.0806437958 |
- |
- |
PREDICTED: putative G3BP-like protein [Jatropha curcas] |
| 7 |
Hb_039946_050 |
0.0816639172 |
- |
- |
catalytic, putative [Ricinus communis] |
| 8 |
Hb_008948_140 |
0.0819108457 |
- |
- |
PREDICTED: nuclear pore complex protein NUP205 [Jatropha curcas] |
| 9 |
Hb_001258_110 |
0.0830734261 |
- |
- |
transporter, putative [Ricinus communis] |
| 10 |
Hb_015026_130 |
0.0832674152 |
- |
- |
quinone oxidoreductase, putative [Ricinus communis] |
| 11 |
Hb_002259_080 |
0.0833676668 |
- |
- |
protein transport protein sec23, putative [Ricinus communis] |
| 12 |
Hb_000635_030 |
0.0837194921 |
- |
- |
mannosyl-oligosaccharide glucosidase, putative [Ricinus communis] |
| 13 |
Hb_007747_170 |
0.0858508112 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 [Jatropha curcas] |
| 14 |
Hb_012286_040 |
0.0860338133 |
- |
- |
PREDICTED: pre-mRNA-splicing factor RSE1 [Jatropha curcas] |
| 15 |
Hb_000001_170 |
0.0869760389 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 16 |
Hb_000580_050 |
0.0904128214 |
- |
- |
- |
| 17 |
Hb_019840_030 |
0.0906819054 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X2 [Jatropha curcas] |
| 18 |
Hb_007657_020 |
0.0917629441 |
- |
- |
PREDICTED: PRA1 family protein H isoform X1 [Jatropha curcas] |
| 19 |
Hb_000130_020 |
0.0919654335 |
- |
- |
Phosphoenolpyruvate carboxylase, putative [Ricinus communis] |
| 20 |
Hb_000260_780 |
0.0921072954 |
- |
- |
PREDICTED: ERAD-associated E3 ubiquitin-protein ligase HRD1B-like [Jatropha curcas] |