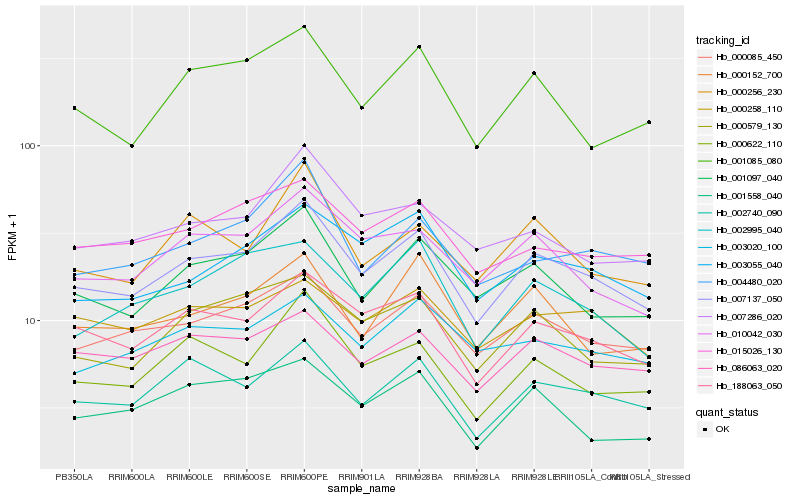
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007137_050 |
0.0 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 2 |
Hb_188063_050 |
0.0635267269 |
- |
- |
PREDICTED: putative G3BP-like protein [Jatropha curcas] |
| 3 |
Hb_000085_450 |
0.066790113 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 20 [Jatropha curcas] |
| 4 |
Hb_001097_040 |
0.0745813097 |
- |
- |
PREDICTED: probable dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 3B [Jatropha curcas] |
| 5 |
Hb_010042_030 |
0.0778021487 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine diphosphorylase 1 [Jatropha curcas] |
| 6 |
Hb_002740_090 |
0.0779112845 |
- |
- |
PREDICTED: putative membrane-bound O-acyltransferase C24H6.01c [Jatropha curcas] |
| 7 |
Hb_000579_130 |
0.0797576142 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 8 |
Hb_000622_110 |
0.0825923012 |
- |
- |
cmp-sialic acid transporter, putative [Ricinus communis] |
| 9 |
Hb_001085_080 |
0.0828813349 |
- |
- |
eukaryotic translation elongation factor 1-alpha [Hevea brasiliensis] |
| 10 |
Hb_003055_040 |
0.0853066247 |
- |
- |
PREDICTED: secretory carrier-associated membrane protein 3-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_086063_020 |
0.0870296378 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 [Jatropha curcas] |
| 12 |
Hb_015026_130 |
0.0879101115 |
- |
- |
quinone oxidoreductase, putative [Ricinus communis] |
| 13 |
Hb_007286_020 |
0.0905630186 |
- |
- |
hypothetical protein L484_010675 [Morus notabilis] |
| 14 |
Hb_002995_040 |
0.0923022128 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase dyrk1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000152_700 |
0.0926542831 |
- |
- |
PREDICTED: E3 ubiquitin protein ligase RIN2 [Jatropha curcas] |
| 16 |
Hb_000256_230 |
0.092836758 |
- |
- |
PREDICTED: uncharacterized protein LOC105637594 [Jatropha curcas] |
| 17 |
Hb_000258_110 |
0.0930428062 |
- |
- |
PREDICTED: probable apyrase 6 isoform X1 [Jatropha curcas] |
| 18 |
Hb_003020_100 |
0.0930959557 |
- |
- |
PREDICTED: transcription initiation factor IIA large subunit [Jatropha curcas] |
| 19 |
Hb_001558_040 |
0.0936070542 |
- |
- |
PREDICTED: GPI ethanolamine phosphate transferase 2 [Jatropha curcas] |
| 20 |
Hb_004480_020 |
0.0942703055 |
- |
- |
PREDICTED: uncharacterized protein LOC105637166 [Jatropha curcas] |