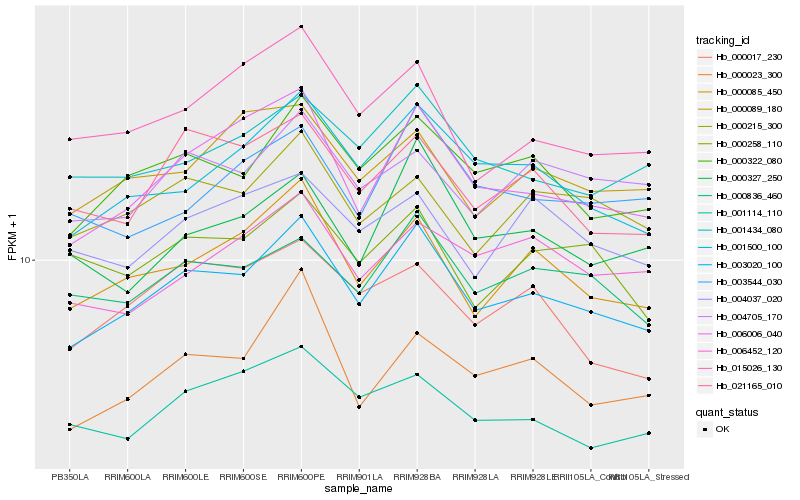
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003020_100 |
0.0 |
- |
- |
PREDICTED: transcription initiation factor IIA large subunit [Jatropha curcas] |
| 2 |
Hb_000322_080 |
0.0557904422 |
- |
- |
hypothetical protein JCGZ_07423 [Jatropha curcas] |
| 3 |
Hb_006006_040 |
0.0574083725 |
- |
- |
S-methyl-5-thioribose kinase isoform 2 [Theobroma cacao] |
| 4 |
Hb_001500_100 |
0.061499711 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000085_450 |
0.0620533044 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 20 [Jatropha curcas] |
| 6 |
Hb_000836_460 |
0.0706961911 |
- |
- |
PREDICTED: uncharacterized protein LOC105628435 [Jatropha curcas] |
| 7 |
Hb_021165_010 |
0.0734619963 |
- |
- |
PREDICTED: splicing factor U2af large subunit B-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_001434_080 |
0.0754351545 |
- |
- |
PREDICTED: probable protein S-acyltransferase 7 [Jatropha curcas] |
| 9 |
Hb_015026_130 |
0.0795792419 |
- |
- |
quinone oxidoreductase, putative [Ricinus communis] |
| 10 |
Hb_000215_300 |
0.0797341619 |
- |
- |
mitochondrial processing peptidase alpha subunit, putative [Ricinus communis] |
| 11 |
Hb_000327_250 |
0.0803271708 |
- |
- |
PREDICTED: macrophage erythroblast attacher [Jatropha curcas] |
| 12 |
Hb_001114_110 |
0.0807532977 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2A isoform X1 [Jatropha curcas] |
| 13 |
Hb_004705_170 |
0.0808484305 |
- |
- |
PREDICTED: ubiquitin thioesterase otubain-like [Jatropha curcas] |
| 14 |
Hb_000089_180 |
0.0821597781 |
- |
- |
PREDICTED: homoserine kinase-like [Jatropha curcas] |
| 15 |
Hb_000023_300 |
0.0823513011 |
- |
- |
Ubiquitin carboxyl-terminal hydrolase, putative [Ricinus communis] |
| 16 |
Hb_003544_030 |
0.0827090572 |
- |
- |
PREDICTED: citrate synthase, glyoxysomal [Jatropha curcas] |
| 17 |
Hb_006452_120 |
0.0848144153 |
- |
- |
PREDICTED: protein FAM188A [Jatropha curcas] |
| 18 |
Hb_000258_110 |
0.0864532812 |
- |
- |
PREDICTED: probable apyrase 6 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000017_230 |
0.0865509537 |
- |
- |
hypothetical protein JCGZ_20558 [Jatropha curcas] |
| 20 |
Hb_004037_020 |
0.0867668969 |
- |
- |
PREDICTED: uncharacterized protein LOC105632366 isoform X1 [Jatropha curcas] |