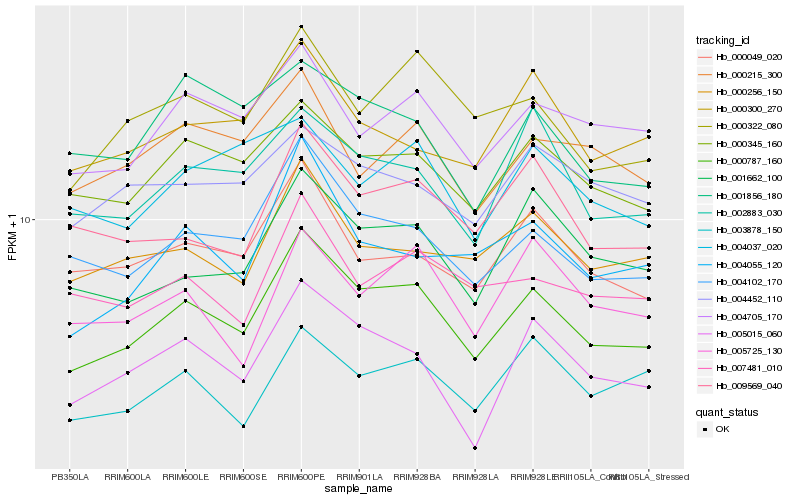
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000787_160 |
0.0 |
- |
- |
PREDICTED: GDP-mannose transporter GONST1 isoform X1 [Jatropha curcas] |
| 2 |
Hb_004102_170 |
0.0567447938 |
- |
- |
PREDICTED: lys-63-specific deubiquitinase BRCC36-like [Jatropha curcas] |
| 3 |
Hb_000345_160 |
0.0634739038 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 4-like [Jatropha curcas] |
| 4 |
Hb_005015_060 |
0.0668859681 |
- |
- |
PREDICTED: developmentally-regulated G-protein 3 [Jatropha curcas] |
| 5 |
Hb_000322_080 |
0.0690363586 |
- |
- |
hypothetical protein JCGZ_07423 [Jatropha curcas] |
| 6 |
Hb_004705_170 |
0.0691077859 |
- |
- |
PREDICTED: ubiquitin thioesterase otubain-like [Jatropha curcas] |
| 7 |
Hb_001662_100 |
0.0692115988 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1-like [Jatropha curcas] |
| 8 |
Hb_002883_030 |
0.0715590989 |
- |
- |
PREDICTED: uncharacterized protein LOC105638222 [Jatropha curcas] |
| 9 |
Hb_000049_020 |
0.0726339298 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 10 |
Hb_009569_040 |
0.0744509364 |
- |
- |
PREDICTED: uncharacterized protein LOC105635573 [Jatropha curcas] |
| 11 |
Hb_004055_120 |
0.0745301786 |
- |
- |
PREDICTED: uncharacterized protein LOC105631443 [Jatropha curcas] |
| 12 |
Hb_000256_150 |
0.0746055174 |
- |
- |
PREDICTED: trafficking protein particle complex II-specific subunit 120 homolog [Jatropha curcas] |
| 13 |
Hb_003878_150 |
0.0777363877 |
- |
- |
PREDICTED: GPI mannosyltransferase 3 [Jatropha curcas] |
| 14 |
Hb_000300_270 |
0.0797359939 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 15 |
Hb_000215_300 |
0.079846754 |
- |
- |
mitochondrial processing peptidase alpha subunit, putative [Ricinus communis] |
| 16 |
Hb_007481_010 |
0.079870263 |
- |
- |
PREDICTED: TBC1 domain family member 22B [Jatropha curcas] |
| 17 |
Hb_004452_110 |
0.0824423805 |
- |
- |
PREDICTED: uncharacterized protein LOC105639575 isoform X1 [Jatropha curcas] |
| 18 |
Hb_005725_130 |
0.0843055665 |
- |
- |
PREDICTED: uncharacterized protein KIAA0930 homolog isoform X2 [Jatropha curcas] |
| 19 |
Hb_001856_180 |
0.0847642869 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit M [Jatropha curcas] |
| 20 |
Hb_004037_020 |
0.0848327485 |
- |
- |
PREDICTED: uncharacterized protein LOC105632366 isoform X1 [Jatropha curcas] |