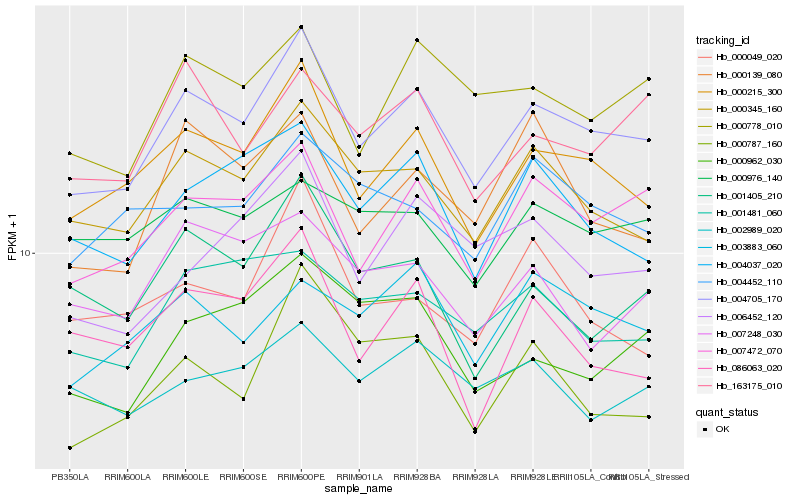
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004705_170 |
0.0 |
- |
- |
PREDICTED: ubiquitin thioesterase otubain-like [Jatropha curcas] |
| 2 |
Hb_000215_300 |
0.0399865892 |
- |
- |
mitochondrial processing peptidase alpha subunit, putative [Ricinus communis] |
| 3 |
Hb_007472_070 |
0.0498816576 |
- |
- |
cir, putative [Ricinus communis] |
| 4 |
Hb_000345_160 |
0.0516698887 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 4-like [Jatropha curcas] |
| 5 |
Hb_004037_020 |
0.0627207543 |
- |
- |
PREDICTED: uncharacterized protein LOC105632366 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000778_010 |
0.0641906424 |
- |
- |
hypothetical protein [Bacillus subtilis] |
| 7 |
Hb_163175_010 |
0.0660708586 |
- |
- |
hypothetical protein CISIN_1g036035mg, partial [Citrus sinensis] |
| 8 |
Hb_001481_060 |
0.0667793218 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HOS1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_003883_060 |
0.0674354009 |
transcription factor |
TF Family: BSD |
PREDICTED: probable RNA polymerase II transcription factor B subunit 1-1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000787_160 |
0.0691077859 |
- |
- |
PREDICTED: GDP-mannose transporter GONST1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000962_030 |
0.0724706562 |
- |
- |
PREDICTED: metal tolerance protein C2 [Jatropha curcas] |
| 12 |
Hb_000139_080 |
0.0724878154 |
- |
- |
PREDICTED: vacuole membrane protein KMS1 isoform X2 [Jatropha curcas] |
| 13 |
Hb_004452_110 |
0.0724968714 |
- |
- |
PREDICTED: uncharacterized protein LOC105639575 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001405_210 |
0.0726808807 |
- |
- |
hypothetical protein JCGZ_22540 [Jatropha curcas] |
| 15 |
Hb_000976_140 |
0.0727788654 |
- |
- |
PREDICTED: uncharacterized protein LOC105646208 isoform X1 [Jatropha curcas] |
| 16 |
Hb_006452_120 |
0.0730077886 |
- |
- |
PREDICTED: protein FAM188A [Jatropha curcas] |
| 17 |
Hb_007248_030 |
0.0734317515 |
- |
- |
PREDICTED: uncharacterized protein LOC105633558 [Jatropha curcas] |
| 18 |
Hb_002989_020 |
0.0736073868 |
- |
- |
PREDICTED: uncharacterized protein LOC105630013 isoform X2 [Jatropha curcas] |
| 19 |
Hb_086063_020 |
0.074579236 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 [Jatropha curcas] |
| 20 |
Hb_000049_020 |
0.0752192677 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |