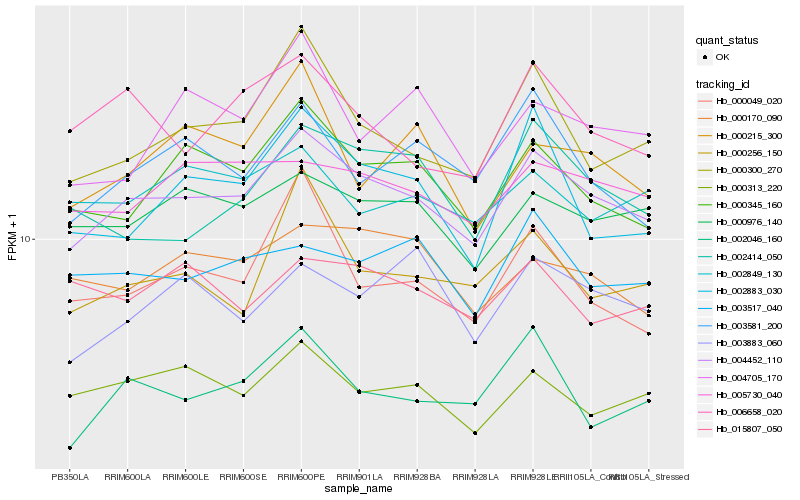
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004452_110 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105639575 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000345_160 |
0.0552728269 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 4-like [Jatropha curcas] |
| 3 |
Hb_000976_140 |
0.0568235849 |
- |
- |
PREDICTED: uncharacterized protein LOC105646208 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000300_270 |
0.058030795 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 5 |
Hb_002046_160 |
0.0587318571 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 6 |
Hb_005730_040 |
0.0600384702 |
- |
- |
hypothetical protein PRUPE_ppa004338mg [Prunus persica] |
| 7 |
Hb_000313_220 |
0.0604904833 |
- |
- |
PREDICTED: sulfhydryl oxidase 2 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000170_090 |
0.0650779736 |
- |
- |
PREDICTED: UDP-sugar pyrophosphorylase [Jatropha curcas] |
| 9 |
Hb_003517_040 |
0.0653617951 |
- |
- |
PREDICTED: putative DUF21 domain-containing protein At3g13070, chloroplastic [Jatropha curcas] |
| 10 |
Hb_006658_020 |
0.0654746229 |
- |
- |
PREDICTED: uncharacterized protein LOC105633529 [Jatropha curcas] |
| 11 |
Hb_003581_200 |
0.0659032351 |
- |
- |
PREDICTED: uncharacterized protein LOC105640179 [Jatropha curcas] |
| 12 |
Hb_000256_150 |
0.0668817809 |
- |
- |
PREDICTED: trafficking protein particle complex II-specific subunit 120 homolog [Jatropha curcas] |
| 13 |
Hb_000049_020 |
0.0676611824 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 14 |
Hb_003883_060 |
0.0687066656 |
transcription factor |
TF Family: BSD |
PREDICTED: probable RNA polymerase II transcription factor B subunit 1-1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002849_130 |
0.0689344176 |
- |
- |
PREDICTED: RAB6A-GEF complex partner protein 1-like [Jatropha curcas] |
| 16 |
Hb_002414_050 |
0.0696141635 |
- |
- |
hypothetical protein CICLE_v10000243mg [Citrus clementina] |
| 17 |
Hb_002883_030 |
0.0699829758 |
- |
- |
PREDICTED: uncharacterized protein LOC105638222 [Jatropha curcas] |
| 18 |
Hb_004705_170 |
0.0724968714 |
- |
- |
PREDICTED: ubiquitin thioesterase otubain-like [Jatropha curcas] |
| 19 |
Hb_000215_300 |
0.0727814095 |
- |
- |
mitochondrial processing peptidase alpha subunit, putative [Ricinus communis] |
| 20 |
Hb_015807_050 |
0.0728084476 |
- |
- |
PREDICTED: uncharacterized protein LOC105632878 [Jatropha curcas] |