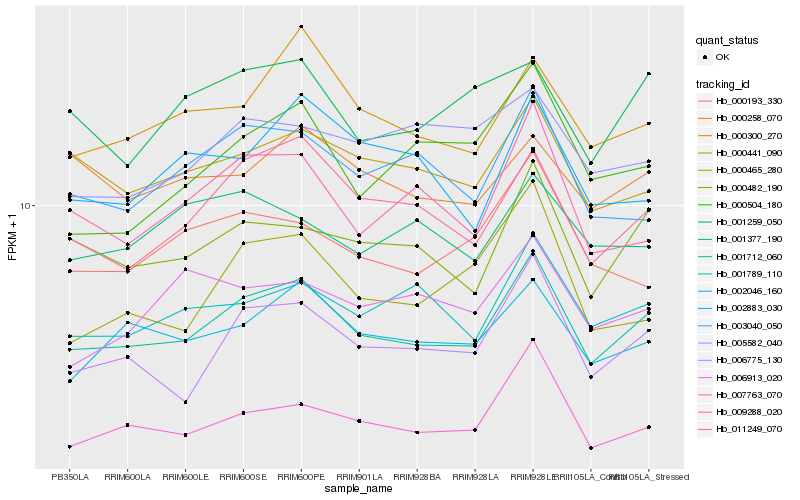
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001712_060 |
0.0 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK9 [Jatropha curcas] |
| 2 |
Hb_006775_130 |
0.0576975061 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET2a-like [Jatropha curcas] |
| 3 |
Hb_011249_070 |
0.0632075706 |
- |
- |
hypothetical protein POPTR_0018s12610g [Populus trichocarpa] |
| 4 |
Hb_000482_190 |
0.0639703507 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 5 |
Hb_007763_070 |
0.0641940878 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g71460, chloroplastic [Jatropha curcas] |
| 6 |
Hb_002046_160 |
0.0646595436 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 7 |
Hb_000300_270 |
0.0686504649 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 8 |
Hb_001789_110 |
0.0686598519 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 9 |
Hb_000441_090 |
0.0691208563 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 6 regulatory subunit 2 [Jatropha curcas] |
| 10 |
Hb_009288_020 |
0.0754568289 |
- |
- |
PREDICTED: protein FRIGIDA [Jatropha curcas] |
| 11 |
Hb_003040_050 |
0.0764673276 |
- |
- |
PREDICTED: uncharacterized protein LOC105638219 [Jatropha curcas] |
| 12 |
Hb_001259_050 |
0.0801160034 |
- |
- |
calcium lipid binding protein, putative [Ricinus communis] |
| 13 |
Hb_000258_070 |
0.0811110166 |
- |
- |
PREDICTED: uncharacterized protein LOC105649104 [Jatropha curcas] |
| 14 |
Hb_000193_330 |
0.0812998858 |
- |
- |
PREDICTED: formin-like protein 20 [Jatropha curcas] |
| 15 |
Hb_000504_180 |
0.0857371268 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 16 |
Hb_002883_030 |
0.086170751 |
- |
- |
PREDICTED: uncharacterized protein LOC105638222 [Jatropha curcas] |
| 17 |
Hb_000465_280 |
0.0864502897 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g47360 [Jatropha curcas] |
| 18 |
Hb_001377_190 |
0.0866616596 |
transcription factor |
TF Family: PHD |
PREDICTED: histone-lysine N-methyltransferase ATX3 isoform X2 [Jatropha curcas] |
| 19 |
Hb_006913_020 |
0.0867572059 |
- |
- |
PREDICTED: uncharacterized protein LOC105649145 isoform X1 [Jatropha curcas] |
| 20 |
Hb_005582_040 |
0.0869246247 |
- |
- |
PREDICTED: regulator of nonsense transcripts 1 homolog [Jatropha curcas] |