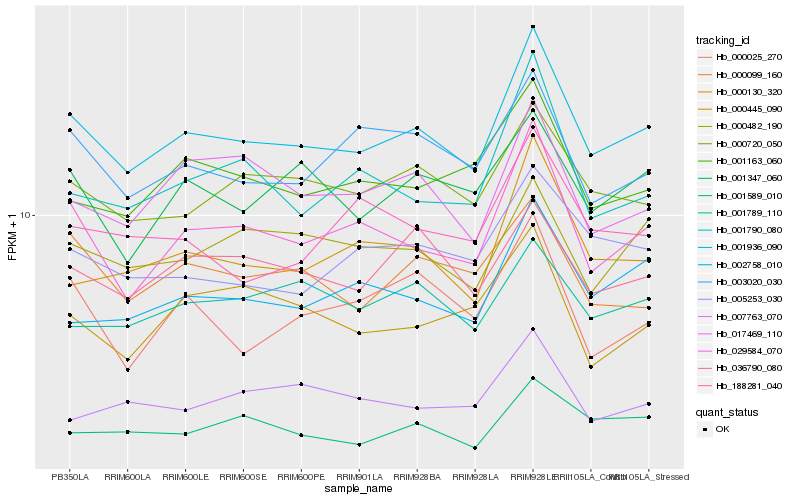
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001936_090 |
0.0 |
- |
- |
PREDICTED: BOI-related E3 ubiquitin-protein ligase 1-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_029584_070 |
0.0605735765 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g15690-like [Jatropha curcas] |
| 3 |
Hb_000720_050 |
0.0706551331 |
- |
- |
PREDICTED: uncharacterized protein LOC105645857 isoform X1 [Jatropha curcas] |
| 4 |
Hb_002758_010 |
0.0787370085 |
- |
- |
PREDICTED: ribonuclease II, chloroplastic/mitochondrial [Populus euphratica] |
| 5 |
Hb_000130_320 |
0.0789644198 |
desease resistance |
Gene Name: AAA |
PREDICTED: uncharacterized protein ycf45 [Jatropha curcas] |
| 6 |
Hb_188281_040 |
0.0793009319 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 12 [Jatropha curcas] |
| 7 |
Hb_001347_060 |
0.0801205122 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 8 |
Hb_003020_030 |
0.0804817879 |
- |
- |
PREDICTED: protein TOC75-3, chloroplastic-like [Populus euphratica] |
| 9 |
Hb_001163_060 |
0.0806171664 |
- |
- |
neutral/alkaline invertase [Manihot esculenta] |
| 10 |
Hb_005253_030 |
0.0809951933 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_001589_010 |
0.0813009552 |
- |
- |
PREDICTED: uncharacterized protein LOC105638192 isoform X1 [Jatropha curcas] |
| 12 |
Hb_017469_110 |
0.0823493389 |
- |
- |
PREDICTED: uncharacterized protein LOC105641795 [Jatropha curcas] |
| 13 |
Hb_000482_190 |
0.082767277 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 14 |
Hb_000099_160 |
0.0835346887 |
- |
- |
PREDICTED: potassium transporter 3 isoform X2 [Populus euphratica] |
| 15 |
Hb_001790_080 |
0.0841363644 |
- |
- |
PREDICTED: chloroplastic group IIA intron splicing facilitator CRS1, chloroplastic isoform X3 [Jatropha curcas] |
| 16 |
Hb_001789_110 |
0.0873844073 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 17 |
Hb_036790_080 |
0.0920502887 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 39 [Jatropha curcas] |
| 18 |
Hb_000445_090 |
0.0939092348 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH2 [Jatropha curcas] |
| 19 |
Hb_000025_270 |
0.0947460409 |
- |
- |
PREDICTED: proteinaceous RNase P 1, chloroplastic/mitochondrial-like [Jatropha curcas] |
| 20 |
Hb_007763_070 |
0.0956908159 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g71460, chloroplastic [Jatropha curcas] |