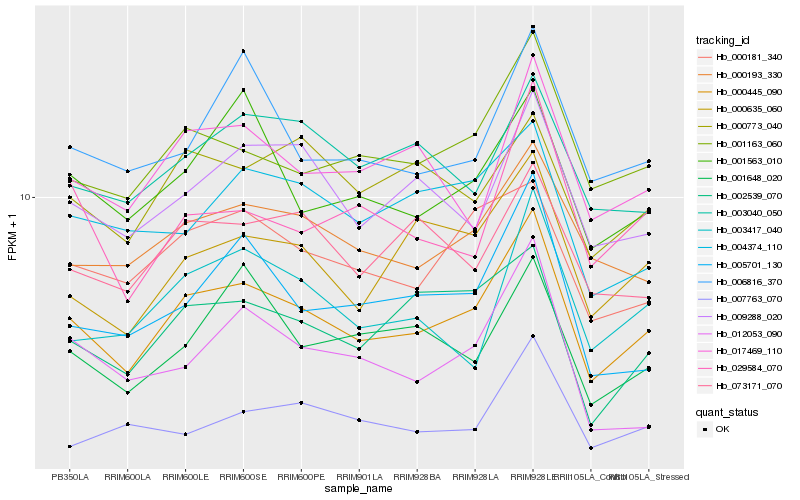
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000445_090 |
0.0 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH2 [Jatropha curcas] |
| 2 |
Hb_001163_060 |
0.0669121156 |
- |
- |
neutral/alkaline invertase [Manihot esculenta] |
| 3 |
Hb_000193_330 |
0.072947148 |
- |
- |
PREDICTED: formin-like protein 20 [Jatropha curcas] |
| 4 |
Hb_000635_060 |
0.0787243474 |
- |
- |
PREDICTED: uncharacterized protein LOC105644797 [Jatropha curcas] |
| 5 |
Hb_000181_340 |
0.0792093603 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 6 |
Hb_009288_020 |
0.0795988686 |
- |
- |
PREDICTED: protein FRIGIDA [Jatropha curcas] |
| 7 |
Hb_002539_070 |
0.082557207 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g26460, mitochondrial [Jatropha curcas] |
| 8 |
Hb_017469_110 |
0.0833557138 |
- |
- |
PREDICTED: uncharacterized protein LOC105641795 [Jatropha curcas] |
| 9 |
Hb_001563_010 |
0.0848422823 |
transcription factor |
TF Family: SET |
PREDICTED: probable histone-lysine N-methyltransferase ATXR3 [Jatropha curcas] |
| 10 |
Hb_006816_370 |
0.0863451346 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105647456 [Jatropha curcas] |
| 11 |
Hb_073171_070 |
0.0875102817 |
- |
- |
PREDICTED: RNA polymerase II-associated factor 1 homolog [Jatropha curcas] |
| 12 |
Hb_005701_130 |
0.087768625 |
transcription factor |
TF Family: SWI/SNF-SWI3 |
PREDICTED: lysine-specific histone demethylase 1 homolog 3 [Jatropha curcas] |
| 13 |
Hb_004374_110 |
0.089495862 |
- |
- |
PREDICTED: uncharacterized protein LOC105645768 [Jatropha curcas] |
| 14 |
Hb_003040_050 |
0.0896581267 |
- |
- |
PREDICTED: uncharacterized protein LOC105638219 [Jatropha curcas] |
| 15 |
Hb_000773_040 |
0.0898815126 |
- |
- |
PREDICTED: uncharacterized protein LOC105641863 isoform X2 [Jatropha curcas] |
| 16 |
Hb_029584_070 |
0.0903872186 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g15690-like [Jatropha curcas] |
| 17 |
Hb_003417_040 |
0.0906861983 |
transcription factor |
TF Family: mTERF |
hypothetical protein JCGZ_10155 [Jatropha curcas] |
| 18 |
Hb_007763_070 |
0.0912491348 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g71460, chloroplastic [Jatropha curcas] |
| 19 |
Hb_001648_020 |
0.0923902929 |
- |
- |
hypothetical protein POPTR_0007s07750g [Populus trichocarpa] |
| 20 |
Hb_012053_090 |
0.0924876839 |
transcription factor |
TF Family: DDT |
PREDICTED: uncharacterized protein LOC105637346 [Jatropha curcas] |