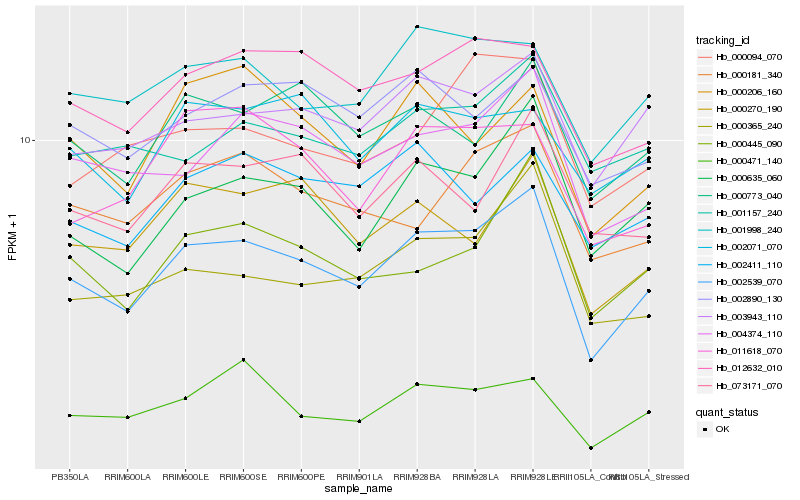
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002539_070 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g26460, mitochondrial [Jatropha curcas] |
| 2 |
Hb_000635_060 |
0.0728478727 |
- |
- |
PREDICTED: uncharacterized protein LOC105644797 [Jatropha curcas] |
| 3 |
Hb_004374_110 |
0.0737929329 |
- |
- |
PREDICTED: uncharacterized protein LOC105645768 [Jatropha curcas] |
| 4 |
Hb_003943_110 |
0.0794515556 |
transcription factor |
TF Family: MYB-related |
Zuotin, putative [Ricinus communis] |
| 5 |
Hb_000445_090 |
0.082557207 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH2 [Jatropha curcas] |
| 6 |
Hb_012632_010 |
0.0834963135 |
- |
- |
PREDICTED: uncharacterized protein LOC105640019 [Jatropha curcas] |
| 7 |
Hb_000365_240 |
0.0839376887 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 33A-like [Jatropha curcas] |
| 8 |
Hb_001998_240 |
0.0841452643 |
- |
- |
PREDICTED: small nuclear ribonucleoprotein-associated protein B' [Jatropha curcas] |
| 9 |
Hb_000773_040 |
0.0876132318 |
- |
- |
PREDICTED: uncharacterized protein LOC105641863 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000270_190 |
0.0881467603 |
transcription factor |
TF Family: DDT |
tRNA ligase, putative [Ricinus communis] |
| 11 |
Hb_002890_130 |
0.0881565569 |
- |
- |
tip120, putative [Ricinus communis] |
| 12 |
Hb_000181_340 |
0.08883605 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 13 |
Hb_073171_070 |
0.0914229742 |
- |
- |
PREDICTED: RNA polymerase II-associated factor 1 homolog [Jatropha curcas] |
| 14 |
Hb_000471_140 |
0.0918284396 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g28640 [Prunus mume] |
| 15 |
Hb_000206_160 |
0.0925029013 |
- |
- |
PREDICTED: TBCC domain-containing protein 1 [Jatropha curcas] |
| 16 |
Hb_000094_070 |
0.0936312215 |
- |
- |
PREDICTED: 5'-nucleotidase domain-containing protein 4 isoform X2 [Jatropha curcas] |
| 17 |
Hb_001157_240 |
0.094039513 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 37-like [Jatropha curcas] |
| 18 |
Hb_011618_070 |
0.0944369081 |
- |
- |
NMDA receptor-regulated protein [Medicago truncatula] |
| 19 |
Hb_002411_110 |
0.0947041908 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit RPC2 [Jatropha curcas] |
| 20 |
Hb_002071_070 |
0.0966624573 |
transcription factor |
TF Family: RB |
conserved hypothetical protein [Ricinus communis] |