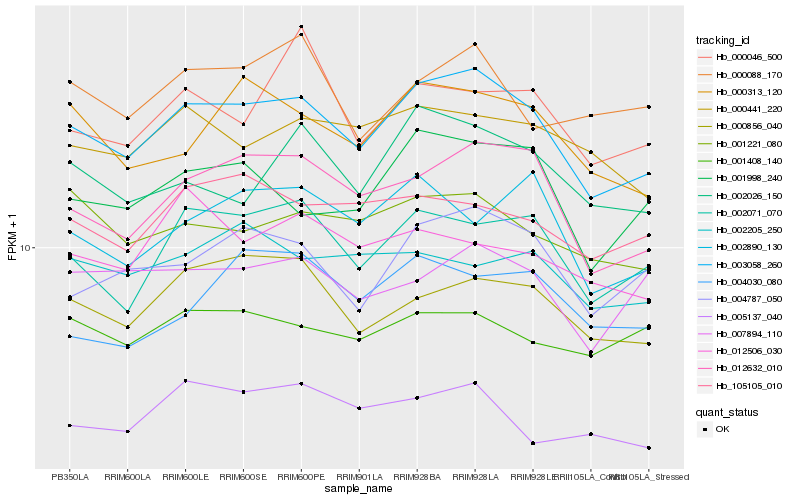
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003058_260 |
0.0 |
- |
- |
hypothetical protein POPTR_0003s13040g [Populus trichocarpa] |
| 2 |
Hb_012632_010 |
0.0503362525 |
- |
- |
PREDICTED: uncharacterized protein LOC105640019 [Jatropha curcas] |
| 3 |
Hb_002071_070 |
0.0595897031 |
transcription factor |
TF Family: RB |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_004787_050 |
0.063628298 |
- |
- |
PREDICTED: RINT1-like protein MAG2 [Jatropha curcas] |
| 5 |
Hb_000856_040 |
0.0645118584 |
transcription factor |
TF Family: DDT |
PREDICTED: uncharacterized protein LOC105640470 [Jatropha curcas] |
| 6 |
Hb_000313_120 |
0.0661953598 |
- |
- |
PREDICTED: ALG-2 interacting protein X [Jatropha curcas] |
| 7 |
Hb_001998_240 |
0.066562022 |
- |
- |
PREDICTED: small nuclear ribonucleoprotein-associated protein B' [Jatropha curcas] |
| 8 |
Hb_001221_080 |
0.0701834718 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 2 isoform X2 [Jatropha curcas] |
| 9 |
Hb_002026_150 |
0.0710863983 |
transcription factor |
TF Family: SNF2 |
PREDICTED: DNA repair helicase XPB1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000441_220 |
0.07261543 |
- |
- |
PREDICTED: histidinol dehydrogenase, chloroplastic isoform X1 [Jatropha curcas] |
| 11 |
Hb_105105_010 |
0.0731440434 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 132 [Jatropha curcas] |
| 12 |
Hb_002890_130 |
0.0747208542 |
- |
- |
tip120, putative [Ricinus communis] |
| 13 |
Hb_005137_040 |
0.0748108776 |
- |
- |
hypothetical protein JCGZ_12324 [Jatropha curcas] |
| 14 |
Hb_007894_110 |
0.0748321839 |
- |
- |
Heat-shock protein 105 kDa, putative [Ricinus communis] |
| 15 |
Hb_001408_140 |
0.0787467821 |
- |
- |
hypothetical protein JCGZ_00234 [Jatropha curcas] |
| 16 |
Hb_000088_170 |
0.079381186 |
- |
- |
PREDICTED: ran-binding protein 10-like [Jatropha curcas] |
| 17 |
Hb_002205_250 |
0.0806329138 |
- |
- |
PREDICTED: probable inactive serine/threonine-protein kinase scy1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000046_500 |
0.0813209113 |
- |
- |
PREDICTED: probable serine/threonine-protein phosphatase 2A regulatory subunit B'' subunit TON2 [Jatropha curcas] |
| 19 |
Hb_012506_030 |
0.0819834879 |
- |
- |
AP-2 complex subunit alpha, putative [Ricinus communis] |
| 20 |
Hb_004030_080 |
0.082116591 |
- |
- |
hypothetical protein JCGZ_25110 [Jatropha curcas] |