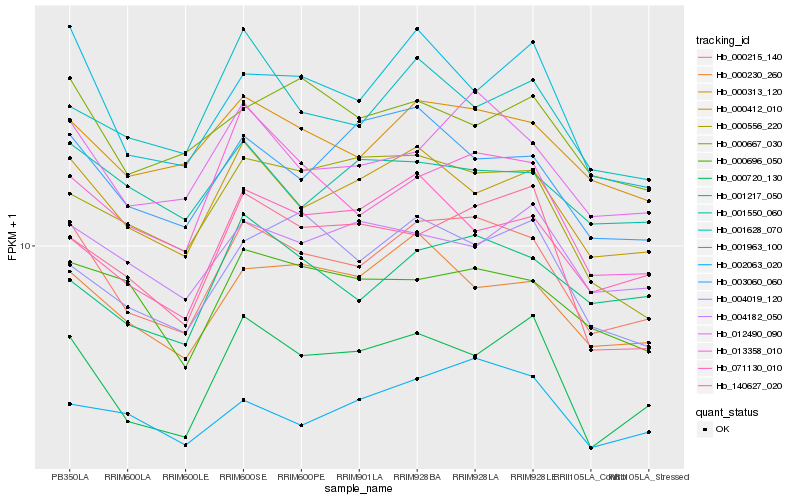
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000215_140 |
0.0 |
- |
- |
PREDICTED: phosphoinositide 3-kinase regulatory subunit 4-like [Malus domestica] |
| 2 |
Hb_012490_090 |
0.0664563178 |
- |
- |
PREDICTED: myosin-17 [Jatropha curcas] |
| 3 |
Hb_000412_010 |
0.0674242214 |
- |
- |
hect ubiquitin-protein ligase, putative [Ricinus communis] |
| 4 |
Hb_000313_120 |
0.0687405673 |
- |
- |
PREDICTED: ALG-2 interacting protein X [Jatropha curcas] |
| 5 |
Hb_000720_130 |
0.0752865699 |
- |
- |
hypothetical protein PRUPE_ppa012392mg [Prunus persica] |
| 6 |
Hb_000230_260 |
0.0754466961 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RF298 [Jatropha curcas] |
| 7 |
Hb_001963_100 |
0.0794883886 |
transcription factor |
TF Family: bZIP |
PREDICTED: bZIP transcription factor 17-like [Jatropha curcas] |
| 8 |
Hb_003060_060 |
0.0836196086 |
- |
- |
PREDICTED: protein transport protein SEC31 homolog B [Jatropha curcas] |
| 9 |
Hb_013358_010 |
0.0836307885 |
- |
- |
hypothetical protein JCGZ_07102 [Jatropha curcas] |
| 10 |
Hb_001217_050 |
0.0843916715 |
- |
- |
actin binding protein, putative [Ricinus communis] |
| 11 |
Hb_071130_010 |
0.0855250978 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000696_050 |
0.0876809732 |
- |
- |
PREDICTED: CSC1-like protein At4g35870 [Jatropha curcas] |
| 13 |
Hb_004019_120 |
0.088346632 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 1 [Jatropha curcas] |
| 14 |
Hb_002063_020 |
0.0892517492 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g17140 [Jatropha curcas] |
| 15 |
Hb_140627_020 |
0.0905619573 |
- |
- |
PREDICTED: uncharacterized protein LOC105637253 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000556_220 |
0.0916414506 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 2-like [Jatropha curcas] |
| 17 |
Hb_001628_070 |
0.0924142074 |
- |
- |
hect ubiquitin-protein ligase, putative [Ricinus communis] |
| 18 |
Hb_000667_030 |
0.092461108 |
- |
- |
PREDICTED: protein transport protein SEC23-like [Jatropha curcas] |
| 19 |
Hb_004182_050 |
0.0929491911 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase BSL3 isoform X2 [Jatropha curcas] |
| 20 |
Hb_001550_060 |
0.0949046543 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase BSL3 isoform X4 [Jatropha curcas] |