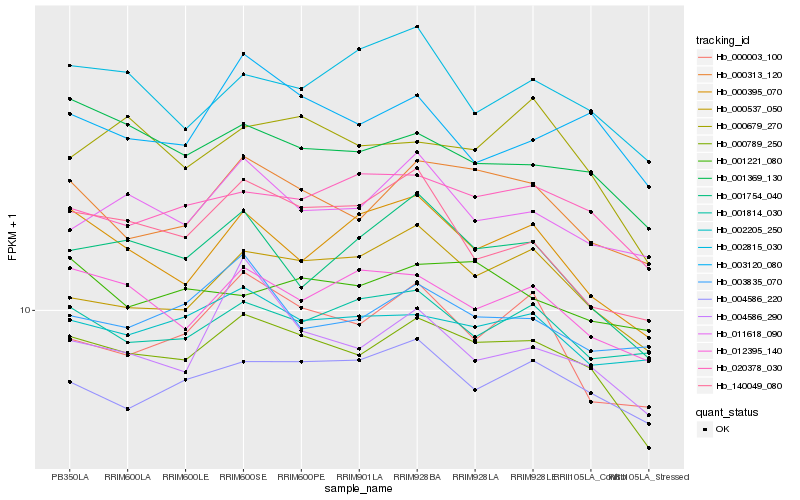
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000789_250 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105637809 [Jatropha curcas] |
| 2 |
Hb_000313_120 |
0.0607560363 |
- |
- |
PREDICTED: ALG-2 interacting protein X [Jatropha curcas] |
| 3 |
Hb_020378_030 |
0.0673129593 |
- |
- |
PREDICTED: protein transport protein SEC31 homolog B [Jatropha curcas] |
| 4 |
Hb_012395_140 |
0.0683643175 |
- |
- |
PREDICTED: uncharacterized protein LOC105638411 [Jatropha curcas] |
| 5 |
Hb_000537_050 |
0.0699545509 |
- |
- |
AP-2 complex subunit beta-1, putative [Ricinus communis] |
| 6 |
Hb_001369_130 |
0.0701274499 |
- |
- |
PREDICTED: autophagy-related protein 3 isoform X1 [Jatropha curcas] |
| 7 |
Hb_004586_220 |
0.0713754566 |
- |
- |
PREDICTED: WD and tetratricopeptide repeats protein 1 isoform X3 [Jatropha curcas] |
| 8 |
Hb_001754_040 |
0.0720071939 |
transcription factor |
TF Family: TRAF |
PREDICTED: uncharacterized protein LOC105643434 [Jatropha curcas] |
| 9 |
Hb_000395_070 |
0.072467412 |
- |
- |
PREDICTED: serine/threonine-protein kinase BLUS1 isoform X2 [Jatropha curcas] |
| 10 |
Hb_002205_250 |
0.0729506481 |
- |
- |
PREDICTED: probable inactive serine/threonine-protein kinase scy1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000679_270 |
0.0740845961 |
- |
- |
PREDICTED: putative clathrin assembly protein At2g25430 [Jatropha curcas] |
| 12 |
Hb_011618_090 |
0.0762154499 |
desease resistance |
Gene Name: ABC_tran |
ABC transporter family protein [Hevea brasiliensis] |
| 13 |
Hb_004586_290 |
0.0788052547 |
- |
- |
PREDICTED: transport inhibitor response 1-like protein [Jatropha curcas] |
| 14 |
Hb_000003_100 |
0.0795386018 |
transcription factor |
TF Family: SNF2 |
PREDICTED: transcription termination factor 2 isoform X5 [Jatropha curcas] |
| 15 |
Hb_003120_080 |
0.0804291472 |
- |
- |
PREDICTED: COPII coat assembly protein SEC16 isoform X1 [Jatropha curcas] |
| 16 |
Hb_140049_080 |
0.0804396243 |
- |
- |
PREDICTED: probable RNA helicase SDE3 [Jatropha curcas] |
| 17 |
Hb_001814_030 |
0.0810477703 |
- |
- |
PREDICTED: uncharacterized protein LOC105650634 [Jatropha curcas] |
| 18 |
Hb_002815_030 |
0.0824864273 |
- |
- |
hypothetical protein CISIN_1g0095162mg, partial [Citrus sinensis] |
| 19 |
Hb_003835_070 |
0.0826426268 |
- |
- |
PREDICTED: uncharacterized protein LOC105641698 isoform X2 [Jatropha curcas] |
| 20 |
Hb_001221_080 |
0.0827972853 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 2 isoform X2 [Jatropha curcas] |