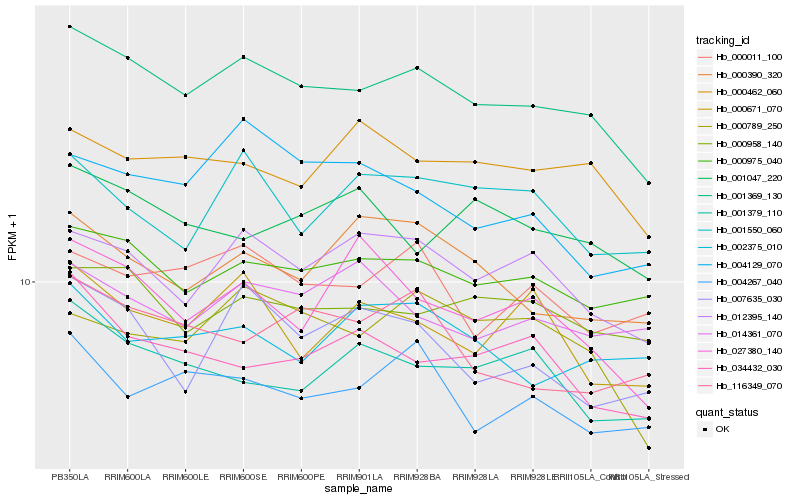
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001369_130 |
0.0 |
- |
- |
PREDICTED: autophagy-related protein 3 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000975_040 |
0.055617454 |
- |
- |
hypothetical protein CICLE_v10028249mg [Citrus clementina] |
| 3 |
Hb_012395_140 |
0.0584885233 |
- |
- |
PREDICTED: uncharacterized protein LOC105638411 [Jatropha curcas] |
| 4 |
Hb_000958_140 |
0.0614583466 |
- |
- |
PREDICTED: uncharacterized protein LOC105628714 [Jatropha curcas] |
| 5 |
Hb_000462_060 |
0.0698572888 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105642316 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000789_250 |
0.0701274499 |
- |
- |
PREDICTED: uncharacterized protein LOC105637809 [Jatropha curcas] |
| 7 |
Hb_004267_040 |
0.0712097219 |
- |
- |
PREDICTED: transcriptional adapter ADA2a [Jatropha curcas] |
| 8 |
Hb_000011_100 |
0.0712907715 |
- |
- |
PREDICTED: uncharacterized protein C630.12 [Jatropha curcas] |
| 9 |
Hb_000390_320 |
0.073655435 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-1b-like [Jatropha curcas] |
| 10 |
Hb_034432_030 |
0.0741831486 |
- |
- |
PREDICTED: probable glycosyltransferase At5g20260 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001379_110 |
0.0742803529 |
- |
- |
PREDICTED: sec1 family domain-containing protein MIP3 [Jatropha curcas] |
| 12 |
Hb_000671_070 |
0.0751538806 |
- |
- |
PREDICTED: uncharacterized protein LOC105630621 [Jatropha curcas] |
| 13 |
Hb_007635_030 |
0.07600942 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g77360, mitochondrial-like [Jatropha curcas] |
| 14 |
Hb_014361_070 |
0.0761849271 |
- |
- |
PREDICTED: molybdate-anion transporter [Jatropha curcas] |
| 15 |
Hb_116349_070 |
0.0768520854 |
- |
- |
hypothetical protein JCGZ_21753 [Jatropha curcas] |
| 16 |
Hb_001047_220 |
0.0770875524 |
- |
- |
PREDICTED: F-box protein SKIP17-like [Jatropha curcas] |
| 17 |
Hb_004129_070 |
0.0773322381 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 5 [Jatropha curcas] |
| 18 |
Hb_001550_060 |
0.0778826131 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase BSL3 isoform X4 [Jatropha curcas] |
| 19 |
Hb_002375_010 |
0.0778932657 |
- |
- |
PREDICTED: C2 and GRAM domain-containing protein At1g03370 isoform X1 [Jatropha curcas] |
| 20 |
Hb_027380_140 |
0.0786118231 |
- |
- |
PREDICTED: uncharacterized protein LOC105634023 isoform X1 [Jatropha curcas] |