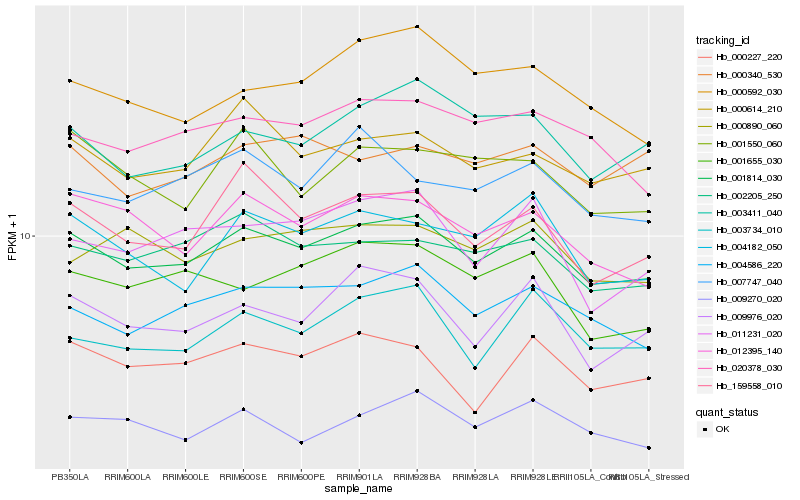
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001814_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105650634 [Jatropha curcas] |
| 2 |
Hb_004586_220 |
0.0447491036 |
- |
- |
PREDICTED: WD and tetratricopeptide repeats protein 1 isoform X3 [Jatropha curcas] |
| 3 |
Hb_002205_250 |
0.0491245134 |
- |
- |
PREDICTED: probable inactive serine/threonine-protein kinase scy1 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000614_210 |
0.050051169 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-1-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_012395_140 |
0.0524139115 |
- |
- |
PREDICTED: uncharacterized protein LOC105638411 [Jatropha curcas] |
| 6 |
Hb_159558_010 |
0.0526037068 |
- |
- |
PREDICTED: uncharacterized protein LOC105635846 [Jatropha curcas] |
| 7 |
Hb_020378_030 |
0.0532155375 |
- |
- |
PREDICTED: protein transport protein SEC31 homolog B [Jatropha curcas] |
| 8 |
Hb_009270_020 |
0.0541996782 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: helicase and polymerase-containing protein TEBICHI [Jatropha curcas] |
| 9 |
Hb_009976_020 |
0.0549219171 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105631286 [Jatropha curcas] |
| 10 |
Hb_004182_050 |
0.0561587105 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase BSL3 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000227_220 |
0.0564960999 |
- |
- |
PREDICTED: protein phosphatase 2C 70 [Jatropha curcas] |
| 12 |
Hb_003734_010 |
0.0566257913 |
- |
- |
PREDICTED: double-strand break repair protein MRE11 [Jatropha curcas] |
| 13 |
Hb_003411_040 |
0.0569680972 |
- |
- |
unnamed protein product [Coffea canephora] |
| 14 |
Hb_001550_060 |
0.0602570536 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase BSL3 isoform X4 [Jatropha curcas] |
| 15 |
Hb_000890_060 |
0.0602705978 |
- |
- |
PREDICTED: sucrose nonfermenting 4-like protein [Jatropha curcas] |
| 16 |
Hb_011231_020 |
0.0609781316 |
- |
- |
PREDICTED: proline-, glutamic acid- and leucine-rich protein 1 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000592_030 |
0.0612608587 |
- |
- |
PREDICTED: uncharacterized protein LOC100853969 isoform X1 [Vitis vinifera] |
| 18 |
Hb_001655_030 |
0.0616059134 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SNF2 domain-containing protein CLASSY 3 isoform X2 [Jatropha curcas] |
| 19 |
Hb_007747_040 |
0.0616640821 |
- |
- |
PREDICTED: probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY [Jatropha curcas] |
| 20 |
Hb_000340_530 |
0.0617447988 |
- |
- |
hypothetical protein VITISV_016664 [Vitis vinifera] |