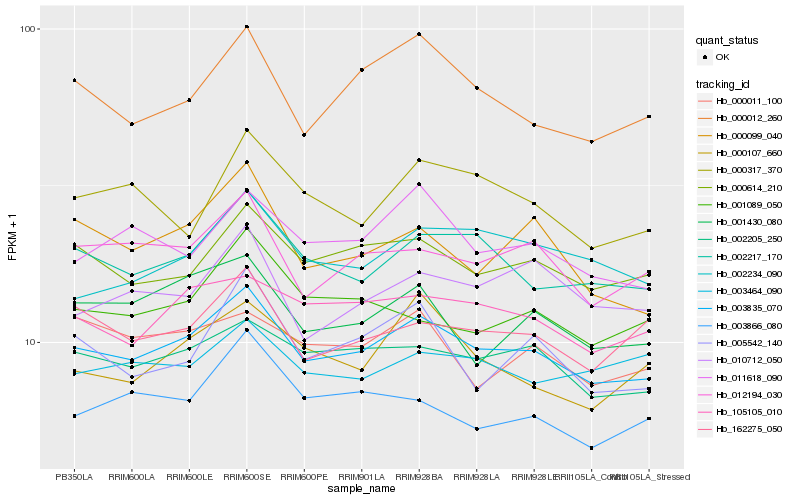
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003835_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105641698 isoform X2 [Jatropha curcas] |
| 2 |
Hb_002205_250 |
0.0471879431 |
- |
- |
PREDICTED: probable inactive serine/threonine-protein kinase scy1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_002217_170 |
0.0480327013 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL6-like isoform X1 [Gossypium raimondii] |
| 4 |
Hb_000614_210 |
0.0486594363 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-1-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_105105_010 |
0.0533957149 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 132 [Jatropha curcas] |
| 6 |
Hb_012194_030 |
0.0544274597 |
- |
- |
PREDICTED: R3H domain-containing protein 2-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_011618_090 |
0.0576438553 |
desease resistance |
Gene Name: ABC_tran |
ABC transporter family protein [Hevea brasiliensis] |
| 8 |
Hb_162275_050 |
0.0595302748 |
- |
- |
PREDICTED: mRNA-capping enzyme [Jatropha curcas] |
| 9 |
Hb_000012_260 |
0.0595559886 |
- |
- |
Far upstream element-binding 2 [Gossypium arboreum] |
| 10 |
Hb_001430_080 |
0.0611688942 |
- |
- |
PREDICTED: putative mediator of RNA polymerase II transcription subunit 26 [Jatropha curcas] |
| 11 |
Hb_002234_090 |
0.0630083833 |
- |
- |
PREDICTED: probable transcriptional regulator SLK2 [Jatropha curcas] |
| 12 |
Hb_003866_080 |
0.0637357842 |
- |
- |
PREDICTED: uncharacterized protein LOC105639150 [Jatropha curcas] |
| 13 |
Hb_010712_050 |
0.0650141298 |
- |
- |
PREDICTED: protein ELC-like [Jatropha curcas] |
| 14 |
Hb_000107_660 |
0.0651921019 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 [Jatropha curcas] |
| 15 |
Hb_005542_140 |
0.0652356448 |
- |
- |
PREDICTED: U-box domain-containing protein 62-like [Jatropha curcas] |
| 16 |
Hb_000099_040 |
0.0655101018 |
- |
- |
unnamed protein product [Coffea canephora] |
| 17 |
Hb_001089_050 |
0.0662506882 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 4 [Jatropha curcas] |
| 18 |
Hb_003464_090 |
0.0665615132 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105642342 [Jatropha curcas] |
| 19 |
Hb_000317_370 |
0.0667836563 |
transcription factor |
TF Family: PHD |
PREDICTED: PHD finger protein At1g33420-like [Jatropha curcas] |
| 20 |
Hb_000011_100 |
0.0670724907 |
- |
- |
PREDICTED: uncharacterized protein C630.12 [Jatropha curcas] |