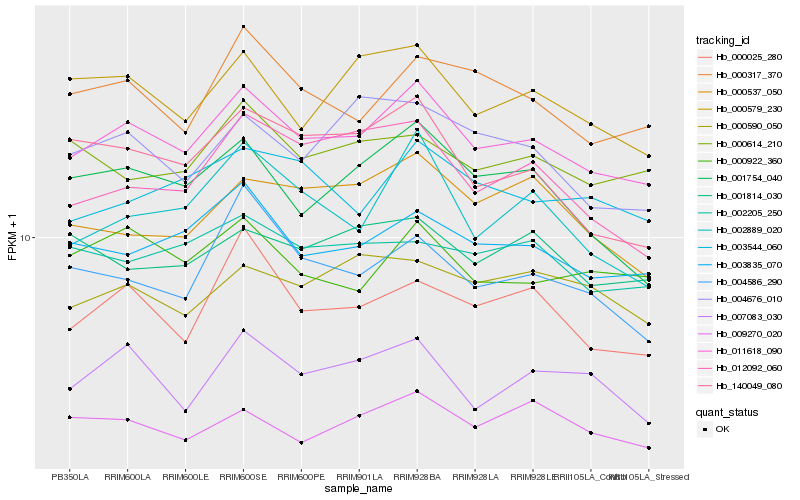
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011618_090 |
0.0 |
desease resistance |
Gene Name: ABC_tran |
ABC transporter family protein [Hevea brasiliensis] |
| 2 |
Hb_003835_070 |
0.0576438553 |
- |
- |
PREDICTED: uncharacterized protein LOC105641698 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000317_370 |
0.0624433778 |
transcription factor |
TF Family: PHD |
PREDICTED: PHD finger protein At1g33420-like [Jatropha curcas] |
| 4 |
Hb_000579_230 |
0.0647136964 |
- |
- |
PREDICTED: bifunctional nuclease 1 [Jatropha curcas] |
| 5 |
Hb_000922_360 |
0.0671248865 |
- |
- |
- |
| 6 |
Hb_003544_060 |
0.0674207962 |
- |
- |
hypothetical protein JCGZ_22100 [Jatropha curcas] |
| 7 |
Hb_007083_030 |
0.0677120773 |
- |
- |
Ubiquitin carboxyl-terminal hydrolase, putative [Ricinus communis] |
| 8 |
Hb_001754_040 |
0.0682526285 |
transcription factor |
TF Family: TRAF |
PREDICTED: uncharacterized protein LOC105643434 [Jatropha curcas] |
| 9 |
Hb_000025_280 |
0.0686627697 |
transcription factor |
TF Family: C3H |
hypothetical protein JCGZ_07432 [Jatropha curcas] |
| 10 |
Hb_002889_020 |
0.0701869344 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 3 [Jatropha curcas] |
| 11 |
Hb_009270_020 |
0.070857152 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: helicase and polymerase-containing protein TEBICHI [Jatropha curcas] |
| 12 |
Hb_002205_250 |
0.0715780339 |
- |
- |
PREDICTED: probable inactive serine/threonine-protein kinase scy1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_012092_060 |
0.0726082516 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 [Jatropha curcas] |
| 14 |
Hb_140049_080 |
0.0733151272 |
- |
- |
PREDICTED: probable RNA helicase SDE3 [Jatropha curcas] |
| 15 |
Hb_000590_050 |
0.0734427197 |
- |
- |
PREDICTED: protein-tyrosine-phosphatase MKP1 [Jatropha curcas] |
| 16 |
Hb_000614_210 |
0.07356369 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-1-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_004676_010 |
0.073834187 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g80270, mitochondrial-like [Jatropha curcas] |
| 18 |
Hb_004586_290 |
0.0741115864 |
- |
- |
PREDICTED: transport inhibitor response 1-like protein [Jatropha curcas] |
| 19 |
Hb_000537_050 |
0.0742399394 |
- |
- |
AP-2 complex subunit beta-1, putative [Ricinus communis] |
| 20 |
Hb_001814_030 |
0.0748033914 |
- |
- |
PREDICTED: uncharacterized protein LOC105650634 [Jatropha curcas] |