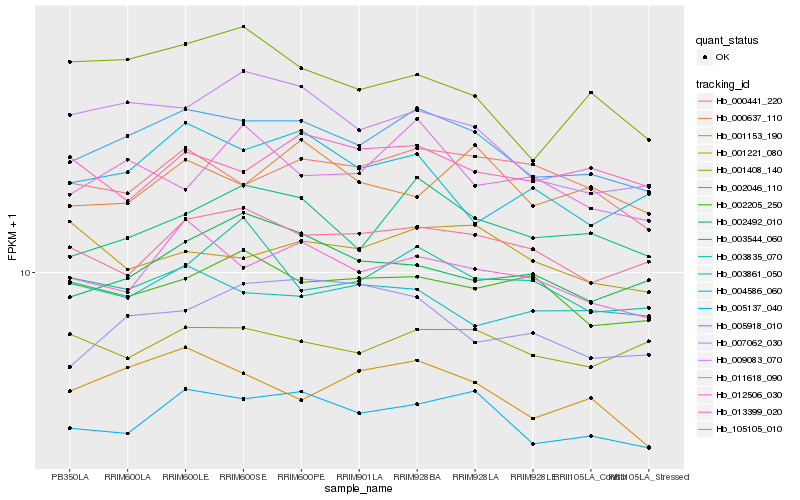
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005918_010 |
0.0 |
- |
- |
PREDICTED: protein ROOT HAIR DEFECTIVE 3-like [Jatropha curcas] |
| 2 |
Hb_005137_040 |
0.0463715185 |
- |
- |
hypothetical protein JCGZ_12324 [Jatropha curcas] |
| 3 |
Hb_012506_030 |
0.0564342899 |
- |
- |
AP-2 complex subunit alpha, putative [Ricinus communis] |
| 4 |
Hb_003544_060 |
0.058379135 |
- |
- |
hypothetical protein JCGZ_22100 [Jatropha curcas] |
| 5 |
Hb_001153_190 |
0.0622506055 |
- |
- |
PREDICTED: DNA-directed RNA polymerase IV subunit 1 [Jatropha curcas] |
| 6 |
Hb_105105_010 |
0.0652803021 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 132 [Jatropha curcas] |
| 7 |
Hb_000441_220 |
0.0654786084 |
- |
- |
PREDICTED: histidinol dehydrogenase, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_009083_070 |
0.0710633036 |
- |
- |
PREDICTED: uncharacterized protein LOC105639870 [Jatropha curcas] |
| 9 |
Hb_001408_140 |
0.0721987576 |
- |
- |
hypothetical protein JCGZ_00234 [Jatropha curcas] |
| 10 |
Hb_002205_250 |
0.0740393414 |
- |
- |
PREDICTED: probable inactive serine/threonine-protein kinase scy1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002492_010 |
0.0767486693 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 2-like [Populus euphratica] |
| 12 |
Hb_004586_060 |
0.077012855 |
- |
- |
PREDICTED: AP-4 complex subunit mu [Jatropha curcas] |
| 13 |
Hb_003861_050 |
0.0770135192 |
- |
- |
PREDICTED: uncharacterized protein LOC105650779 [Jatropha curcas] |
| 14 |
Hb_003835_070 |
0.0782825892 |
- |
- |
PREDICTED: uncharacterized protein LOC105641698 isoform X2 [Jatropha curcas] |
| 15 |
Hb_011618_090 |
0.0794378766 |
desease resistance |
Gene Name: ABC_tran |
ABC transporter family protein [Hevea brasiliensis] |
| 16 |
Hb_013399_020 |
0.0797237505 |
- |
- |
PREDICTED: developmentally-regulated G-protein 2 [Jatropha curcas] |
| 17 |
Hb_002046_110 |
0.079774769 |
- |
- |
PREDICTED: TBC1 domain family member 15 isoform X2 [Jatropha curcas] |
| 18 |
Hb_007062_030 |
0.080827257 |
- |
- |
hypothetical protein VITISV_039033 [Vitis vinifera] |
| 19 |
Hb_000637_110 |
0.0809794555 |
- |
- |
hypothetical protein B456_013G081300 [Gossypium raimondii] |
| 20 |
Hb_001221_080 |
0.0813091047 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 2 isoform X2 [Jatropha curcas] |