| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
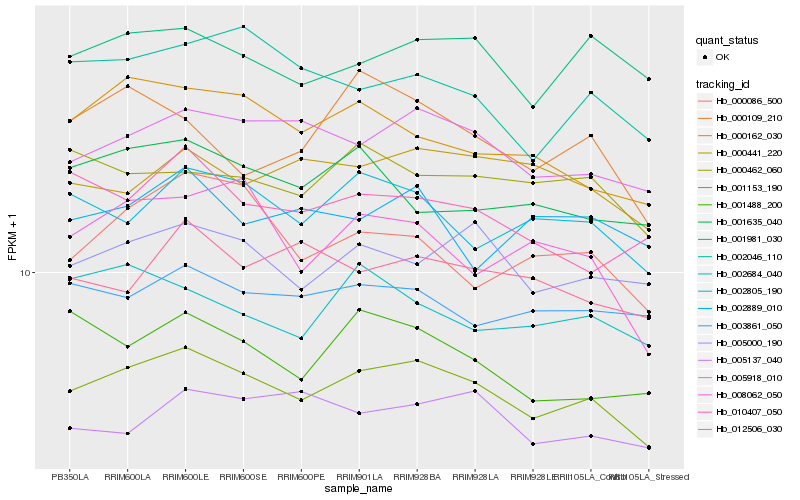
Hb_001153_190 |
0.0 |
- |
- |
PREDICTED: DNA-directed RNA polymerase IV subunit 1 [Jatropha curcas] |
| 2 |
Hb_005918_010 |
0.0622506055 |
- |
- |
PREDICTED: protein ROOT HAIR DEFECTIVE 3-like [Jatropha curcas] |
| 3 |
Hb_005000_190 |
0.0732777402 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 64 [Jatropha curcas] |
| 4 |
Hb_002805_190 |
0.0755149307 |
- |
- |
spliceosome associated protein, putative [Ricinus communis] |
| 5 |
Hb_001981_030 |
0.0765611392 |
- |
- |
nuclear acid binding protein, putative [Ricinus communis] |
| 6 |
Hb_000162_030 |
0.0785449656 |
desease resistance |
Gene Name: RLI |
ABC transporter family protein [Hevea brasiliensis] |
| 7 |
Hb_002046_110 |
0.0816145732 |
- |
- |
PREDICTED: TBC1 domain family member 15 isoform X2 [Jatropha curcas] |
| 8 |
Hb_003861_050 |
0.082690966 |
- |
- |
PREDICTED: uncharacterized protein LOC105650779 [Jatropha curcas] |
| 9 |
Hb_000462_060 |
0.083880335 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105642316 isoform X2 [Jatropha curcas] |
| 10 |
Hb_001488_200 |
0.0851909661 |
- |
- |
PREDICTED: ribosome biogenesis protein WDR12 homolog isoform X1 [Jatropha curcas] |
| 11 |
Hb_010407_050 |
0.0856593859 |
- |
- |
PREDICTED: serine/threonine-protein kinase tricorner-like [Jatropha curcas] |
| 12 |
Hb_000086_500 |
0.0859268197 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 13 |
Hb_000441_220 |
0.086132833 |
- |
- |
PREDICTED: histidinol dehydrogenase, chloroplastic isoform X1 [Jatropha curcas] |
| 14 |
Hb_012506_030 |
0.0867141522 |
- |
- |
AP-2 complex subunit alpha, putative [Ricinus communis] |
| 15 |
Hb_008062_050 |
0.0867213379 |
- |
- |
PREDICTED: uncharacterized protein LOC105631631 [Jatropha curcas] |
| 16 |
Hb_005137_040 |
0.0879011362 |
- |
- |
hypothetical protein JCGZ_12324 [Jatropha curcas] |
| 17 |
Hb_001635_040 |
0.0879027851 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 55 kDa regulatory subunit B beta isoform-like isoform X2 [Gossypium raimondii] |
| 18 |
Hb_002684_040 |
0.0890832096 |
- |
- |
PREDICTED: uncharacterized protein LOC105634392 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000109_210 |
0.0908883981 |
- |
- |
PREDICTED: threonine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 20 |
Hb_002889_010 |
0.0921472208 |
- |
- |
ubx domain-containing, putative [Ricinus communis] |