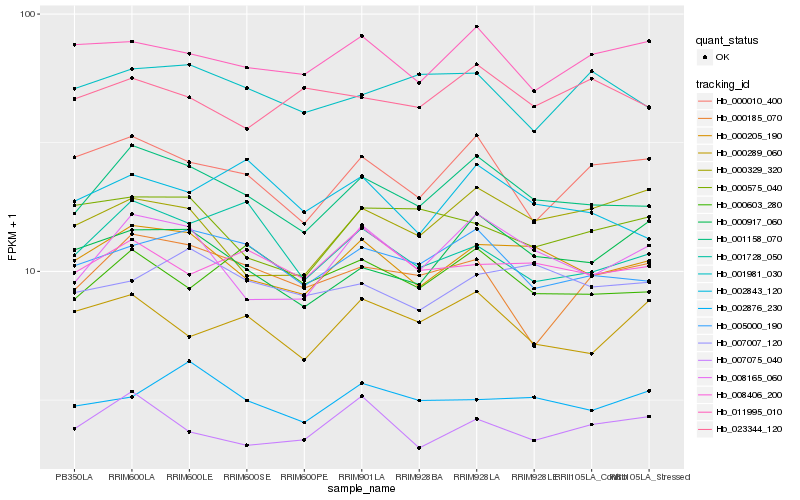
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001158_070 |
0.0 |
- |
- |
Protein DEK, putative [Ricinus communis] |
| 2 |
Hb_000205_190 |
0.055679294 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 3 |
Hb_005000_190 |
0.0617262267 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 64 [Jatropha curcas] |
| 4 |
Hb_008165_060 |
0.0624426599 |
- |
- |
PREDICTED: uncharacterized protein LOC105635541 [Jatropha curcas] |
| 5 |
Hb_000010_400 |
0.0789689133 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 67 [Jatropha curcas] |
| 6 |
Hb_000603_280 |
0.0806945157 |
- |
- |
PREDICTED: uncharacterized protein LOC105638082 [Jatropha curcas] |
| 7 |
Hb_002843_120 |
0.0847876958 |
- |
- |
PREDICTED: uncharacterized protein LOC105645113 isoform X1 [Jatropha curcas] |
| 8 |
Hb_011995_010 |
0.0858569767 |
- |
- |
dc50, putative [Ricinus communis] |
| 9 |
Hb_000917_060 |
0.0860000284 |
- |
- |
PREDICTED: GTP-binding protein At2g22870 [Jatropha curcas] |
| 10 |
Hb_001981_030 |
0.0864864116 |
- |
- |
nuclear acid binding protein, putative [Ricinus communis] |
| 11 |
Hb_001728_050 |
0.0878203832 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RBBP6-like [Jatropha curcas] |
| 12 |
Hb_008406_200 |
0.0879960429 |
transcription factor |
TF Family: SNF2 |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000185_070 |
0.0897226773 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_007007_120 |
0.0898280228 |
- |
- |
PREDICTED: lysophospholipid acyltransferase LPEAT2 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002876_230 |
0.0902820901 |
- |
- |
PREDICTED: uncharacterized protein LOC105633933 [Jatropha curcas] |
| 16 |
Hb_000289_060 |
0.0915920029 |
- |
- |
PREDICTED: uncharacterized protein At1g18480 [Jatropha curcas] |
| 17 |
Hb_007075_040 |
0.0927519562 |
- |
- |
hypothetical protein CISIN_1g039518mg, partial [Citrus sinensis] |
| 18 |
Hb_000575_040 |
0.0931249617 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor SC35 [Jatropha curcas] |
| 19 |
Hb_023344_120 |
0.0936196268 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 9, mitochondrial [Jatropha curcas] |
| 20 |
Hb_000329_320 |
0.0955632188 |
- |
- |
PREDICTED: DNA ligase 1 [Jatropha curcas] |