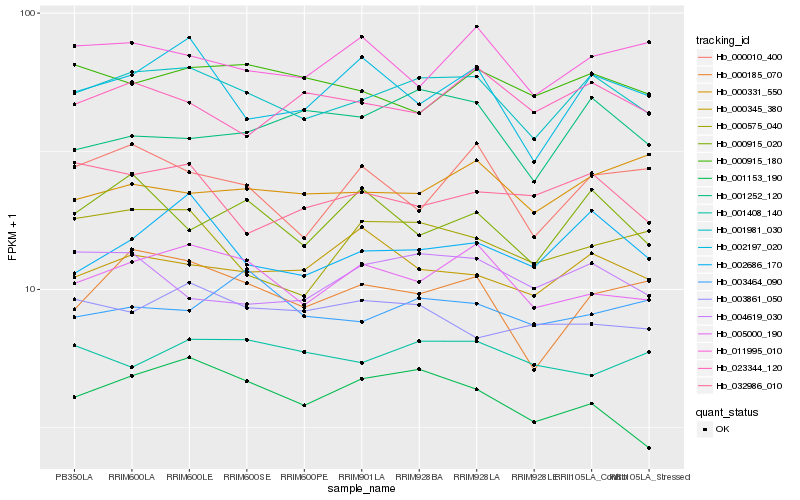
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001981_030 |
0.0 |
- |
- |
nuclear acid binding protein, putative [Ricinus communis] |
| 2 |
Hb_005000_190 |
0.0623159507 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 64 [Jatropha curcas] |
| 3 |
Hb_002686_170 |
0.0686507606 |
- |
- |
PREDICTED: uncharacterized protein LOC105632645 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000185_070 |
0.071675609 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001153_190 |
0.0765611392 |
- |
- |
PREDICTED: DNA-directed RNA polymerase IV subunit 1 [Jatropha curcas] |
| 6 |
Hb_002197_020 |
0.0768340137 |
- |
- |
DNA-directed RNA polymerase family protein [Theobroma cacao] |
| 7 |
Hb_000915_180 |
0.0779614341 |
- |
- |
PREDICTED: V-type proton ATPase 16 kDa proteolipid subunit-like [Pyrus x bretschneideri] |
| 8 |
Hb_023344_120 |
0.0782475062 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 9, mitochondrial [Jatropha curcas] |
| 9 |
Hb_000345_380 |
0.0783068061 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 7, peroxisomal [Jatropha curcas] |
| 10 |
Hb_004619_030 |
0.0795302713 |
- |
- |
PREDICTED: uncharacterized protein LOC105633364 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000575_040 |
0.0801645484 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor SC35 [Jatropha curcas] |
| 12 |
Hb_032986_010 |
0.0802764413 |
- |
- |
Nicotinate-nucleotide pyrophosphorylase [carboxylating], putative [Ricinus communis] |
| 13 |
Hb_011995_010 |
0.0808014801 |
- |
- |
dc50, putative [Ricinus communis] |
| 14 |
Hb_000915_020 |
0.081305431 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 15 |
Hb_003861_050 |
0.0823690642 |
- |
- |
PREDICTED: uncharacterized protein LOC105650779 [Jatropha curcas] |
| 16 |
Hb_001252_120 |
0.0827954793 |
- |
- |
PREDICTED: delta-1-pyrroline-5-carboxylate synthase-like [Gossypium raimondii] |
| 17 |
Hb_001408_140 |
0.0829771185 |
- |
- |
hypothetical protein JCGZ_00234 [Jatropha curcas] |
| 18 |
Hb_000331_550 |
0.0835179433 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 67 [Jatropha curcas] |
| 19 |
Hb_000010_400 |
0.0840273913 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 67 [Jatropha curcas] |
| 20 |
Hb_003464_090 |
0.0843399691 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105642342 [Jatropha curcas] |