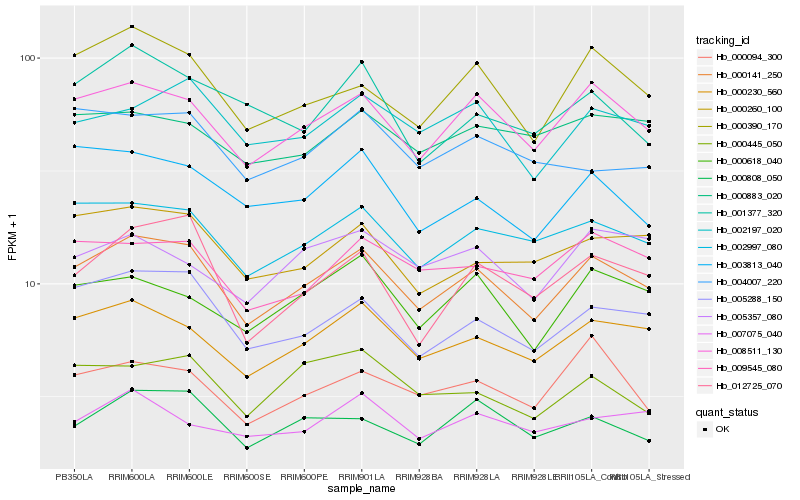
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000141_250 |
0.0 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 2A isoform X3 [Jatropha curcas] |
| 2 |
Hb_008511_130 |
0.0460381787 |
- |
- |
PREDICTED: probable phenylalanine--tRNA ligase alpha subunit [Jatropha curcas] |
| 3 |
Hb_000230_560 |
0.0545652397 |
- |
- |
PREDICTED: UPF0613 protein PB24D3.06c [Jatropha curcas] |
| 4 |
Hb_005288_150 |
0.0571875477 |
- |
- |
PREDICTED: casein kinase I-like isoform X3 [Jatropha curcas] |
| 5 |
Hb_002997_080 |
0.0616382125 |
- |
- |
PREDICTED: uncharacterized protein LOC105631115 [Jatropha curcas] |
| 6 |
Hb_000390_170 |
0.0652364502 |
- |
- |
PREDICTED: V-type proton ATPase subunit H [Jatropha curcas] |
| 7 |
Hb_002197_020 |
0.0724208519 |
- |
- |
DNA-directed RNA polymerase family protein [Theobroma cacao] |
| 8 |
Hb_000260_100 |
0.0746071009 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000808_050 |
0.0752102277 |
- |
- |
exonuclease, putative [Ricinus communis] |
| 10 |
Hb_012725_070 |
0.0759760959 |
- |
- |
PREDICTED: uncharacterized protein LOC105636534 isoform X1 [Jatropha curcas] |
| 11 |
Hb_009545_080 |
0.0765911117 |
- |
- |
hypothetical protein POPTR_0008s15400g [Populus trichocarpa] |
| 12 |
Hb_000618_040 |
0.0776422625 |
- |
- |
hypothetical protein CICLE_v10027942mg [Citrus clementina] |
| 13 |
Hb_003813_040 |
0.0780570156 |
- |
- |
PREDICTED: elongator complex protein 5 [Jatropha curcas] |
| 14 |
Hb_000094_300 |
0.0849367325 |
- |
- |
hypothetical protein POPTR_0002s26360g [Populus trichocarpa] |
| 15 |
Hb_000445_050 |
0.0852622331 |
- |
- |
PREDICTED: DNA topoisomerase 2-binding protein 1 [Jatropha curcas] |
| 16 |
Hb_000883_020 |
0.0868772506 |
- |
- |
PREDICTED: casein kinase II subunit beta-like isoform X5 [Jatropha curcas] |
| 17 |
Hb_007075_040 |
0.0873935002 |
- |
- |
hypothetical protein CISIN_1g039518mg, partial [Citrus sinensis] |
| 18 |
Hb_004007_220 |
0.088720213 |
- |
- |
PREDICTED: casein kinase II subunit alpha [Jatropha curcas] |
| 19 |
Hb_005357_080 |
0.089104938 |
- |
- |
PREDICTED: tyrosyl-DNA phosphodiesterase 1 [Jatropha curcas] |
| 20 |
Hb_001377_320 |
0.0895838772 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH13-like [Jatropha curcas] |