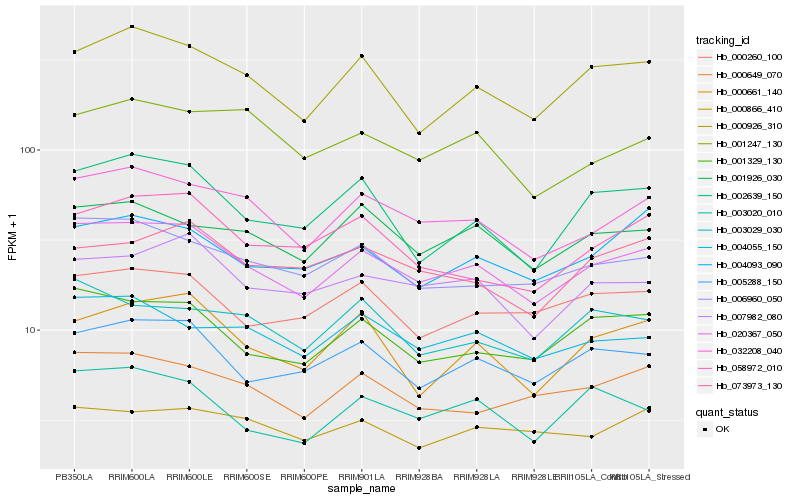
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_020367_050 |
0.0 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranylgeranyl pyrophosphate synthase, putative [Ricinus communis] |
| 2 |
Hb_032208_040 |
0.0532197929 |
- |
- |
PREDICTED: serine/arginine-rich SC35-like splicing factor SCL33 [Populus euphratica] |
| 3 |
Hb_001329_130 |
0.0631354458 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_005288_150 |
0.0648077747 |
- |
- |
PREDICTED: casein kinase I-like isoform X3 [Jatropha curcas] |
| 5 |
Hb_000926_310 |
0.0648678263 |
- |
- |
uncharacterized protein LOC100500241 [Glycine max] |
| 6 |
Hb_006960_050 |
0.0663716083 |
- |
- |
PREDICTED: PRKR-interacting protein 1 [Jatropha curcas] |
| 7 |
Hb_007982_080 |
0.0710533973 |
- |
- |
PREDICTED: uncharacterized protein LOC105636531 [Jatropha curcas] |
| 8 |
Hb_000649_070 |
0.0722146119 |
- |
- |
PREDICTED: probable UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase 2 [Jatropha curcas] |
| 9 |
Hb_004055_150 |
0.0749114668 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002639_150 |
0.0753671896 |
- |
- |
- |
| 11 |
Hb_001926_030 |
0.0755780941 |
- |
- |
PREDICTED: apoptosis inhibitor 5 [Jatropha curcas] |
| 12 |
Hb_000260_100 |
0.0771045266 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_004093_090 |
0.081510083 |
- |
- |
PREDICTED: uncharacterized protein LOC105633550 [Jatropha curcas] |
| 14 |
Hb_058972_010 |
0.081545822 |
- |
- |
PREDICTED: uncharacterized protein LOC105644404 isoform X2 [Jatropha curcas] |
| 15 |
Hb_003020_010 |
0.0822925444 |
transcription factor |
TF Family: G2-like |
PREDICTED: transcription factor LUX [Jatropha curcas] |
| 16 |
Hb_000661_140 |
0.0827130666 |
- |
- |
PREDICTED: uncharacterized protein LOC105648317 isoform X2 [Jatropha curcas] |
| 17 |
Hb_003029_030 |
0.0836373393 |
- |
- |
PREDICTED: uncharacterized protein LOC105642632 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000866_410 |
0.0838138291 |
- |
- |
hypothetical protein JCGZ_14422 [Jatropha curcas] |
| 19 |
Hb_001247_130 |
0.0846346669 |
- |
- |
PREDICTED: shaggy-related protein kinase alpha [Jatropha curcas] |
| 20 |
Hb_073973_130 |
0.0857265374 |
- |
- |
PREDICTED: prolyl-tRNA synthetase associated domain-containing protein 1 [Jatropha curcas] |