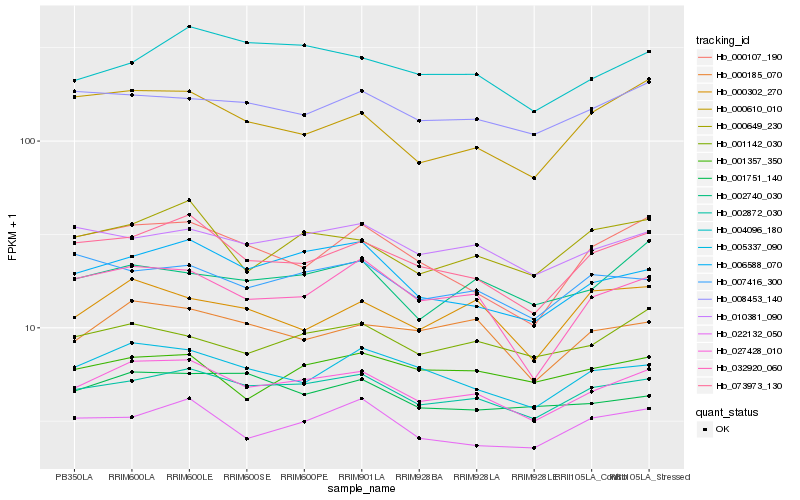
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_027428_010 |
0.0 |
- |
- |
alpha-1,3-mannosyltransferase, putative [Ricinus communis] |
| 2 |
Hb_002872_030 |
0.0414312833 |
- |
- |
PREDICTED: OTU domain-containing protein DDB_G0284757 [Jatropha curcas] |
| 3 |
Hb_073973_130 |
0.0560231291 |
- |
- |
PREDICTED: prolyl-tRNA synthetase associated domain-containing protein 1 [Jatropha curcas] |
| 4 |
Hb_005337_090 |
0.0626408161 |
- |
- |
PREDICTED: serine/threonine-protein kinase TOUSLED-like [Jatropha curcas] |
| 5 |
Hb_006588_070 |
0.0631632618 |
- |
- |
20 kD nuclear cap binding protein, putative [Ricinus communis] |
| 6 |
Hb_000185_070 |
0.0666687867 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_010381_090 |
0.0668266489 |
- |
- |
PREDICTED: protein FLX-like 3 [Jatropha curcas] |
| 8 |
Hb_008453_140 |
0.0702114725 |
- |
- |
PREDICTED: SKP1-like protein 1B [Jatropha curcas] |
| 9 |
Hb_000649_230 |
0.0702489013 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 variant 1A-like [Jatropha curcas] |
| 10 |
Hb_001142_030 |
0.0724998361 |
- |
- |
PREDICTED: uncharacterized protein LOC105640023 [Jatropha curcas] |
| 11 |
Hb_022132_050 |
0.0730086974 |
- |
- |
GTP binding protein, putative [Ricinus communis] |
| 12 |
Hb_032920_060 |
0.0736264314 |
- |
- |
PREDICTED: mitotic checkpoint protein BUB3.1 [Jatropha curcas] |
| 13 |
Hb_004096_180 |
0.0742233278 |
- |
- |
PREDICTED: 14-3-3-like protein A [Jatropha curcas] |
| 14 |
Hb_002740_030 |
0.0754776074 |
transcription factor |
TF Family: bHLH |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001751_140 |
0.0756470334 |
- |
- |
PREDICTED: uncharacterized protein LOC105648510 isoform X1 [Jatropha curcas] |
| 16 |
Hb_007416_300 |
0.0782729722 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 5b-like [Jatropha curcas] |
| 17 |
Hb_001357_350 |
0.0810514283 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000302_270 |
0.0814152851 |
transcription factor |
TF Family: MYB-related |
DNA binding protein, putative [Ricinus communis] |
| 19 |
Hb_000610_010 |
0.0830057453 |
- |
- |
PREDICTED: SKP1-like protein 1B [Musa acuminata subsp. malaccensis] |
| 20 |
Hb_000107_190 |
0.0832253094 |
- |
- |
50S ribosomal protein L20 [Hevea brasiliensis] |