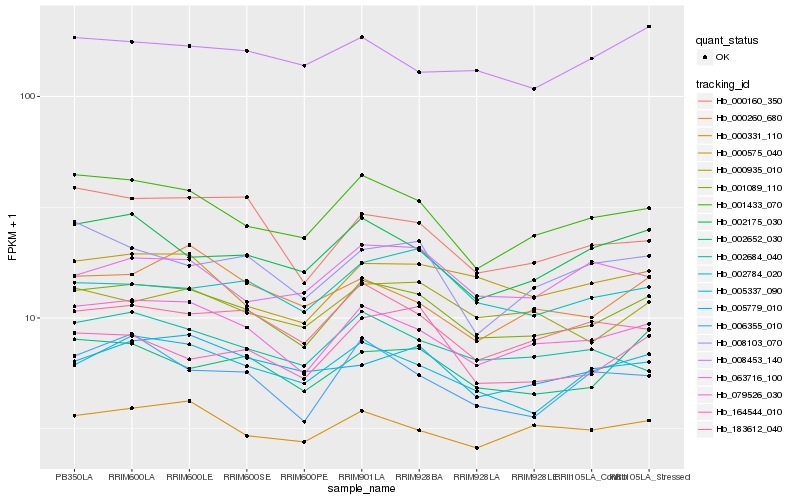
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001089_110 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_079526_030 |
0.0383811324 |
- |
- |
PREDICTED: uncharacterized protein LOC105638120 [Jatropha curcas] |
| 3 |
Hb_001433_070 |
0.0525852567 |
- |
- |
PREDICTED: splicing regulatory glutamine/lysine-rich protein 1 [Jatropha curcas] |
| 4 |
Hb_005337_090 |
0.0576195926 |
- |
- |
PREDICTED: serine/threonine-protein kinase TOUSLED-like [Jatropha curcas] |
| 5 |
Hb_183612_040 |
0.0594251364 |
- |
- |
PREDICTED: uncharacterized protein LOC105632370 [Jatropha curcas] |
| 6 |
Hb_000575_040 |
0.0630787064 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor SC35 [Jatropha curcas] |
| 7 |
Hb_000331_110 |
0.063378136 |
- |
- |
Conserved oligomeric Golgi complex component, putative [Ricinus communis] |
| 8 |
Hb_000935_010 |
0.0637377146 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 16 [Jatropha curcas] |
| 9 |
Hb_002652_030 |
0.0638318074 |
- |
- |
PREDICTED: U3 small nucleolar RNA-interacting protein 2-like [Jatropha curcas] |
| 10 |
Hb_002175_030 |
0.0640989079 |
- |
- |
PREDICTED: snRNA-activating protein complex subunit isoform X2 [Jatropha curcas] |
| 11 |
Hb_006355_010 |
0.0691397105 |
- |
- |
PREDICTED: protein NLRC3 [Jatropha curcas] |
| 12 |
Hb_005779_010 |
0.0698488254 |
- |
- |
catalytic, putative [Ricinus communis] |
| 13 |
Hb_002684_040 |
0.0712213622 |
- |
- |
PREDICTED: uncharacterized protein LOC105634392 isoform X1 [Jatropha curcas] |
| 14 |
Hb_164544_010 |
0.0719444373 |
- |
- |
PREDICTED: protein arginine N-methyltransferase 1.5 isoform X2 [Jatropha curcas] |
| 15 |
Hb_002784_020 |
0.0721239227 |
- |
- |
PREDICTED: probable protein arginine N-methyltransferase 3 [Jatropha curcas] |
| 16 |
Hb_008453_140 |
0.0743413241 |
- |
- |
PREDICTED: SKP1-like protein 1B [Jatropha curcas] |
| 17 |
Hb_063716_100 |
0.0751596321 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 56-like isoform X2 [Jatropha curcas] |
| 18 |
Hb_008103_070 |
0.0777361894 |
transcription factor |
TF Family: MYB-related |
PREDICTED: telomere repeat-binding factor 4-like [Jatropha curcas] |
| 19 |
Hb_000160_350 |
0.0779113372 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 4 isoform X3 [Vitis vinifera] |
| 20 |
Hb_000260_680 |
0.0781209749 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 4 regulatory subunit 2-A [Jatropha curcas] |