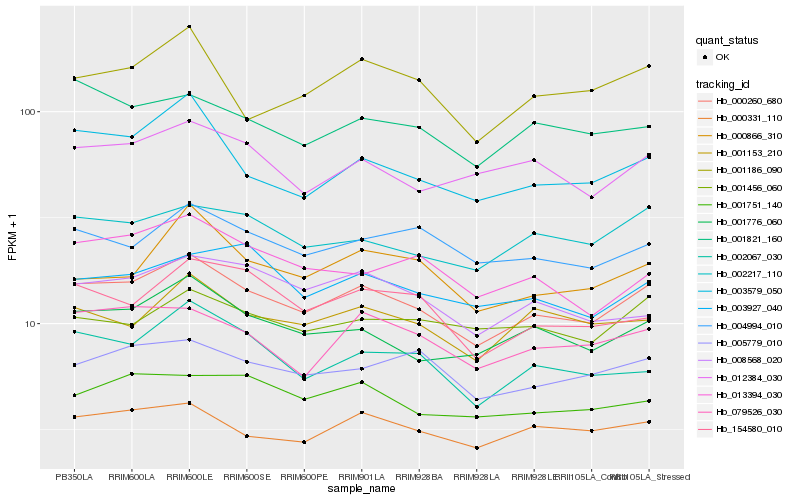
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000260_680 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 4 regulatory subunit 2-A [Jatropha curcas] |
| 2 |
Hb_001776_060 |
0.0564902442 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 5-like [Jatropha curcas] |
| 3 |
Hb_000331_110 |
0.0615222492 |
- |
- |
Conserved oligomeric Golgi complex component, putative [Ricinus communis] |
| 4 |
Hb_002217_110 |
0.0617552829 |
- |
- |
PREDICTED: craniofacial development protein 1 [Jatropha curcas] |
| 5 |
Hb_079526_030 |
0.0664006428 |
- |
- |
PREDICTED: uncharacterized protein LOC105638120 [Jatropha curcas] |
| 6 |
Hb_001153_210 |
0.0666002128 |
- |
- |
PREDICTED: uncharacterized protein LOC105645887 isoform X1 [Jatropha curcas] |
| 7 |
Hb_005779_010 |
0.0673555516 |
- |
- |
catalytic, putative [Ricinus communis] |
| 8 |
Hb_012384_030 |
0.0676293201 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase isozyme L5-like [Jatropha curcas] |
| 9 |
Hb_002067_030 |
0.0685253027 |
- |
- |
PREDICTED: poly(ADP-ribose) glycohydrolase 1-like isoform X2 [Jatropha curcas] |
| 10 |
Hb_004994_010 |
0.0686044546 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 1 [Jatropha curcas] |
| 11 |
Hb_003579_050 |
0.0695891137 |
- |
- |
PREDICTED: 14-3-3 protein 7 [Jatropha curcas] |
| 12 |
Hb_001751_140 |
0.0707067829 |
- |
- |
PREDICTED: uncharacterized protein LOC105648510 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001821_160 |
0.0714024721 |
- |
- |
serine/arginine rich splicing factor, putative [Ricinus communis] |
| 14 |
Hb_008568_020 |
0.0715336348 |
- |
- |
PREDICTED: uncharacterized protein LOC105122325 isoform X3 [Populus euphratica] |
| 15 |
Hb_001186_090 |
0.0722129051 |
- |
- |
PREDICTED: nascent polypeptide-associated complex subunit alpha-like protein 2 [Jatropha curcas] |
| 16 |
Hb_154580_010 |
0.0734059975 |
- |
- |
hypothetical protein PRUPE_ppa002109mg [Prunus persica] |
| 17 |
Hb_003927_040 |
0.0739743248 |
- |
- |
CCR4-NOT transcription complex subunit, putative [Ricinus communis] |
| 18 |
Hb_013394_030 |
0.0742737843 |
- |
- |
PREDICTED: exocyst complex component SEC6 [Vitis vinifera] |
| 19 |
Hb_001456_060 |
0.0743165203 |
- |
- |
hypothetical protein JCGZ_17090 [Jatropha curcas] |
| 20 |
Hb_000866_310 |
0.0744136459 |
- |
- |
PREDICTED: neural Wiskott-Aldrich syndrome protein [Jatropha curcas] |