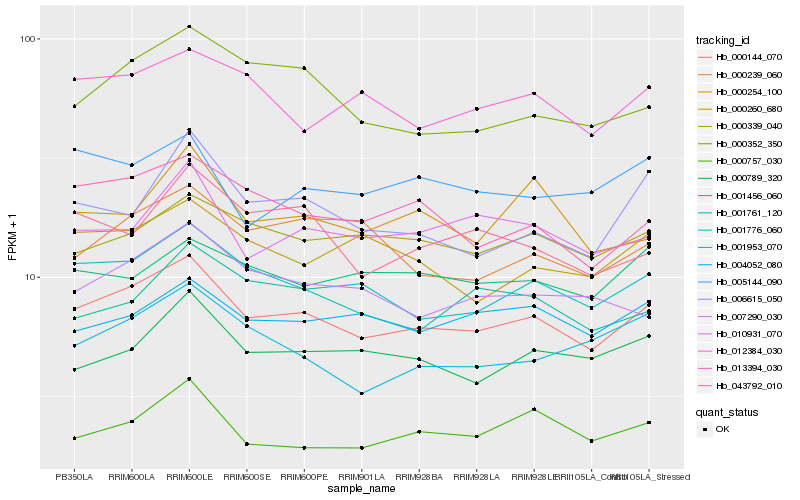
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000144_070 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001776_060 |
0.0584197774 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 5-like [Jatropha curcas] |
| 3 |
Hb_013394_030 |
0.0705185372 |
- |
- |
PREDICTED: exocyst complex component SEC6 [Vitis vinifera] |
| 4 |
Hb_000339_040 |
0.0769031977 |
- |
- |
PREDICTED: SAP30-binding protein isoform X2 [Jatropha curcas] |
| 5 |
Hb_000789_320 |
0.0772840931 |
transcription factor |
TF Family: LUG |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_012384_030 |
0.0777717761 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase isozyme L5-like [Jatropha curcas] |
| 7 |
Hb_000352_350 |
0.0788840026 |
- |
- |
PREDICTED: monothiol glutaredoxin-S7, chloroplastic [Jatropha curcas] |
| 8 |
Hb_010931_070 |
0.0791884628 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 2 isoform X1 [Jatropha curcas] |
| 9 |
Hb_007290_030 |
0.0807896549 |
transcription factor |
TF Family: bZIP |
PREDICTED: uncharacterized protein LOC105629473 [Jatropha curcas] |
| 10 |
Hb_000260_680 |
0.0818554945 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 4 regulatory subunit 2-A [Jatropha curcas] |
| 11 |
Hb_004052_080 |
0.0823199062 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000757_030 |
0.0841265863 |
- |
- |
radical sam protein, putative [Ricinus communis] |
| 13 |
Hb_006615_050 |
0.0842074306 |
- |
- |
PREDICTED: uncharacterized protein LOC105640686 [Jatropha curcas] |
| 14 |
Hb_000239_060 |
0.0857737719 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_005144_090 |
0.086051884 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 24 homolog 1-like [Citrus sinensis] |
| 16 |
Hb_001456_060 |
0.0864109519 |
- |
- |
hypothetical protein JCGZ_17090 [Jatropha curcas] |
| 17 |
Hb_001761_120 |
0.0866475741 |
- |
- |
hypothetical protein JCGZ_22564 [Jatropha curcas] |
| 18 |
Hb_001953_070 |
0.0870530123 |
- |
- |
PREDICTED: RNA pseudouridine synthase 7 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000254_100 |
0.0871015695 |
- |
- |
PREDICTED: glyoxylate/hydroxypyruvate reductase A HPR2 [Jatropha curcas] |
| 20 |
Hb_043792_010 |
0.0874062165 |
- |
- |
PREDICTED: uncharacterized protein LOC105645113 isoform X2 [Jatropha curcas] |