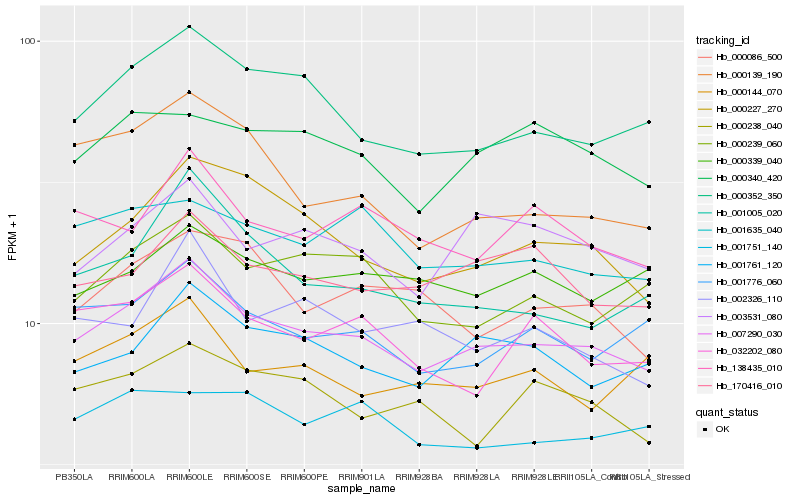
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007290_030 |
0.0 |
transcription factor |
TF Family: bZIP |
PREDICTED: uncharacterized protein LOC105629473 [Jatropha curcas] |
| 2 |
Hb_000352_350 |
0.0658440301 |
- |
- |
PREDICTED: monothiol glutaredoxin-S7, chloroplastic [Jatropha curcas] |
| 3 |
Hb_001761_120 |
0.0700913961 |
- |
- |
hypothetical protein JCGZ_22564 [Jatropha curcas] |
| 4 |
Hb_001776_060 |
0.0716527085 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 5-like [Jatropha curcas] |
| 5 |
Hb_000227_270 |
0.0716734173 |
transcription factor |
TF Family: TRAF |
PREDICTED: ARM REPEAT PROTEIN INTERACTING WITH ABF2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000239_060 |
0.0747628253 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001635_040 |
0.0771378516 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 55 kDa regulatory subunit B beta isoform-like isoform X2 [Gossypium raimondii] |
| 8 |
Hb_032202_080 |
0.0779012506 |
- |
- |
PREDICTED: peroxisome biogenesis protein 16-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_003531_080 |
0.0780069422 |
- |
- |
PREDICTED: small G protein signaling modulator 2 isoform X1 [Jatropha curcas] |
| 10 |
Hb_170416_010 |
0.0785094874 |
- |
- |
PREDICTED: cold-regulated 413 plasma membrane protein 2-like [Jatropha curcas] |
| 11 |
Hb_138435_010 |
0.0788366603 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
brg-1 associated factor, putative [Ricinus communis] |
| 12 |
Hb_000340_420 |
0.0794925876 |
- |
- |
PREDICTED: LEC14B protein isoform X2 [Jatropha curcas] |
| 13 |
Hb_001005_020 |
0.0796787313 |
- |
- |
PREDICTED: uncharacterized protein LOC105630212 [Jatropha curcas] |
| 14 |
Hb_000086_500 |
0.0803113673 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 15 |
Hb_000238_040 |
0.0805752495 |
- |
- |
PREDICTED: uncharacterized protein LOC102611758 [Citrus sinensis] |
| 16 |
Hb_000144_070 |
0.0807896549 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000339_040 |
0.0830424903 |
- |
- |
PREDICTED: SAP30-binding protein isoform X2 [Jatropha curcas] |
| 18 |
Hb_002326_110 |
0.0830828329 |
- |
- |
PREDICTED: uncharacterized protein LOC103320920 [Prunus mume] |
| 19 |
Hb_001751_140 |
0.0833780027 |
- |
- |
PREDICTED: uncharacterized protein LOC105648510 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000139_190 |
0.0847875855 |
transcription factor |
TF Family: TRAF |
PREDICTED: ARM REPEAT PROTEIN INTERACTING WITH ABF2 isoform X2 [Jatropha curcas] |