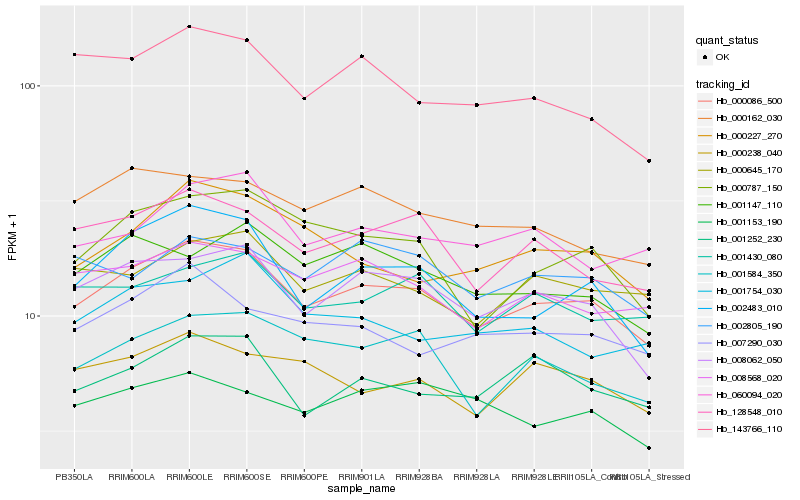
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000086_500 |
0.0 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 2 |
Hb_008062_050 |
0.0611205783 |
- |
- |
PREDICTED: uncharacterized protein LOC105631631 [Jatropha curcas] |
| 3 |
Hb_128548_010 |
0.0676323044 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002483_010 |
0.0683180253 |
transcription factor |
TF Family: IWS1 |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_008568_020 |
0.0724352862 |
- |
- |
PREDICTED: uncharacterized protein LOC105122325 isoform X3 [Populus euphratica] |
| 6 |
Hb_001584_350 |
0.0748403537 |
- |
- |
hypothetical protein JCGZ_23178 [Jatropha curcas] |
| 7 |
Hb_000645_170 |
0.0760445075 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000787_150 |
0.0779604722 |
- |
- |
PREDICTED: protein NDR1-like [Jatropha curcas] |
| 9 |
Hb_001252_230 |
0.0782790416 |
- |
- |
PREDICTED: host cell factor 2 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001430_080 |
0.0788622182 |
- |
- |
PREDICTED: putative mediator of RNA polymerase II transcription subunit 26 [Jatropha curcas] |
| 11 |
Hb_143766_110 |
0.0793442935 |
- |
- |
PREDICTED: asparagine--tRNA ligase, cytoplasmic 1 [Jatropha curcas] |
| 12 |
Hb_000162_030 |
0.0793960116 |
desease resistance |
Gene Name: RLI |
ABC transporter family protein [Hevea brasiliensis] |
| 13 |
Hb_000238_040 |
0.0796289391 |
- |
- |
PREDICTED: uncharacterized protein LOC102611758 [Citrus sinensis] |
| 14 |
Hb_007290_030 |
0.0803113673 |
transcription factor |
TF Family: bZIP |
PREDICTED: uncharacterized protein LOC105629473 [Jatropha curcas] |
| 15 |
Hb_060094_020 |
0.0803262033 |
desease resistance |
Gene Name: ATP_bind_1 |
xpa-binding protein, putative [Ricinus communis] |
| 16 |
Hb_000227_270 |
0.081824186 |
transcription factor |
TF Family: TRAF |
PREDICTED: ARM REPEAT PROTEIN INTERACTING WITH ABF2 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001147_110 |
0.0830298723 |
- |
- |
PREDICTED: dynamin-2A-like [Jatropha curcas] |
| 18 |
Hb_002805_190 |
0.0854438694 |
- |
- |
spliceosome associated protein, putative [Ricinus communis] |
| 19 |
Hb_001754_030 |
0.0856837223 |
- |
- |
PREDICTED: ubiquitin-like-specific protease ESD4 [Jatropha curcas] |
| 20 |
Hb_001153_190 |
0.0859268197 |
- |
- |
PREDICTED: DNA-directed RNA polymerase IV subunit 1 [Jatropha curcas] |