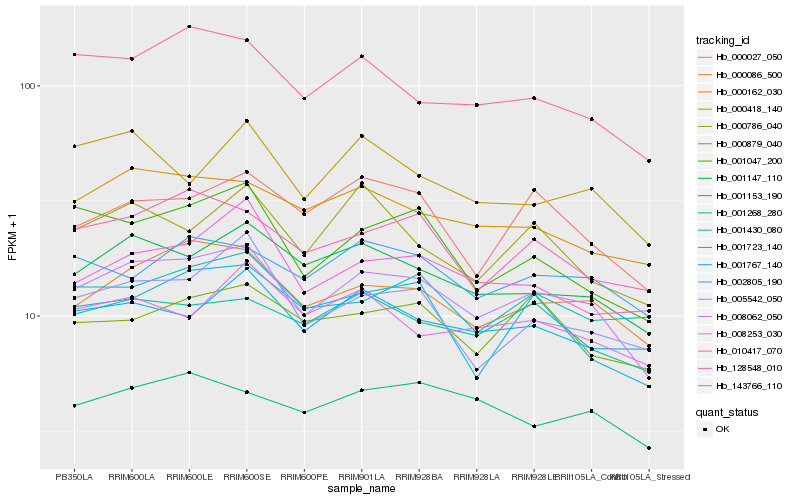
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008062_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631631 [Jatropha curcas] |
| 2 |
Hb_000086_500 |
0.0611205783 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 3 |
Hb_001147_110 |
0.0666290534 |
- |
- |
PREDICTED: dynamin-2A-like [Jatropha curcas] |
| 4 |
Hb_000162_030 |
0.0756619739 |
desease resistance |
Gene Name: RLI |
ABC transporter family protein [Hevea brasiliensis] |
| 5 |
Hb_143766_110 |
0.0759827221 |
- |
- |
PREDICTED: asparagine--tRNA ligase, cytoplasmic 1 [Jatropha curcas] |
| 6 |
Hb_128548_010 |
0.0770013111 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001268_280 |
0.0776023904 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000027_050 |
0.0807255224 |
- |
- |
PREDICTED: uncharacterized protein LOC105636933 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001047_200 |
0.0818901115 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 18 [Eucalyptus grandis] |
| 10 |
Hb_005542_050 |
0.0836734752 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable inactive serine/threonine-protein kinase scy1 [Prunus mume] |
| 11 |
Hb_002805_190 |
0.0844285352 |
- |
- |
spliceosome associated protein, putative [Ricinus communis] |
| 12 |
Hb_001723_140 |
0.0853598431 |
- |
- |
PREDICTED: importin beta-like SAD2 homolog isoform X2 [Jatropha curcas] |
| 13 |
Hb_010417_070 |
0.0859407382 |
- |
- |
PREDICTED: uncharacterized protein LOC105645744 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000879_040 |
0.0864221826 |
- |
- |
PREDICTED: calcium-transporting ATPase 3, endoplasmic reticulum-type [Jatropha curcas] |
| 15 |
Hb_001767_140 |
0.0865915051 |
- |
- |
PREDICTED: protein-tyrosine-phosphatase MKP1 [Jatropha curcas] |
| 16 |
Hb_001153_190 |
0.0867213379 |
- |
- |
PREDICTED: DNA-directed RNA polymerase IV subunit 1 [Jatropha curcas] |
| 17 |
Hb_000786_040 |
0.088531397 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 17 [Jatropha curcas] |
| 18 |
Hb_000418_140 |
0.0888363746 |
- |
- |
PREDICTED: DNA damage-binding protein 1a [Jatropha curcas] |
| 19 |
Hb_001430_080 |
0.0896128468 |
- |
- |
PREDICTED: putative mediator of RNA polymerase II transcription subunit 26 [Jatropha curcas] |
| 20 |
Hb_008253_030 |
0.0896617511 |
- |
- |
PREDICTED: probable carboxylesterase 11 [Jatropha curcas] |