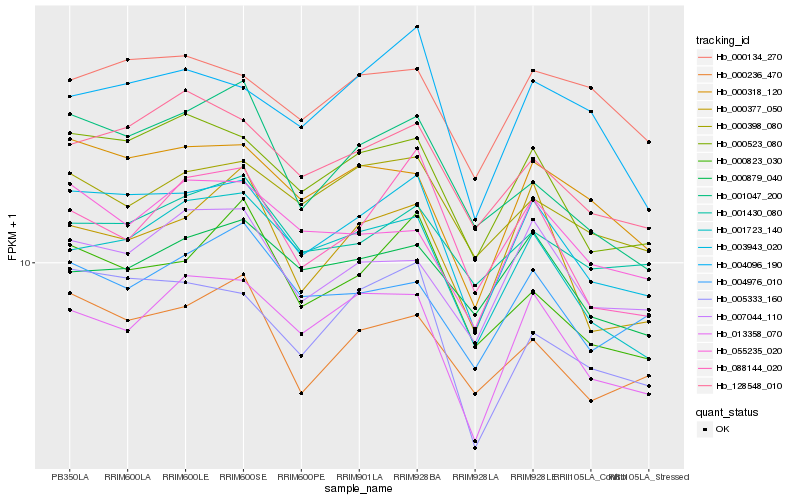
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003943_020 |
0.0 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase PASTICCINO1 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000523_080 |
0.0460042887 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: beta-catenin-like protein 1 [Jatropha curcas] |
| 3 |
Hb_000318_120 |
0.062920331 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: DNA ligase 1 isoform X2 [Jatropha curcas] |
| 4 |
Hb_128548_010 |
0.0652288746 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_005333_160 |
0.0675597704 |
- |
- |
hypothetical protein B456_009G230200 [Gossypium raimondii] |
| 6 |
Hb_013358_070 |
0.0675966655 |
- |
- |
protein with unknown function [Ricinus communis] |
| 7 |
Hb_001723_140 |
0.0679133259 |
- |
- |
PREDICTED: importin beta-like SAD2 homolog isoform X2 [Jatropha curcas] |
| 8 |
Hb_000236_470 |
0.0737387835 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g55840 [Jatropha curcas] |
| 9 |
Hb_007044_110 |
0.0740335647 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 10 |
Hb_000398_080 |
0.0758292305 |
- |
- |
tip120, putative [Ricinus communis] |
| 11 |
Hb_000823_030 |
0.0761466256 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit RPC5 [Jatropha curcas] |
| 12 |
Hb_004096_190 |
0.0762048884 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 13 |
Hb_000377_050 |
0.077220485 |
- |
- |
PREDICTED: sister chromatid cohesion protein PDS5 homolog A isoform X1 [Jatropha curcas] |
| 14 |
Hb_088144_020 |
0.0772225594 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 6-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_055235_020 |
0.0782110267 |
- |
- |
PREDICTED: probable mitochondrial saccharopine dehydrogenase-like oxidoreductase At5g39410 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001430_080 |
0.079202685 |
- |
- |
PREDICTED: putative mediator of RNA polymerase II transcription subunit 26 [Jatropha curcas] |
| 17 |
Hb_001047_200 |
0.0794871369 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 18 [Eucalyptus grandis] |
| 18 |
Hb_004976_010 |
0.0803703969 |
- |
- |
hypothetical protein RCOM_1429110 [Ricinus communis] |
| 19 |
Hb_000879_040 |
0.0805429782 |
- |
- |
PREDICTED: calcium-transporting ATPase 3, endoplasmic reticulum-type [Jatropha curcas] |
| 20 |
Hb_000134_270 |
0.0813951426 |
- |
- |
arginine/serine rich splicing factor sf4/14, putative [Ricinus communis] |