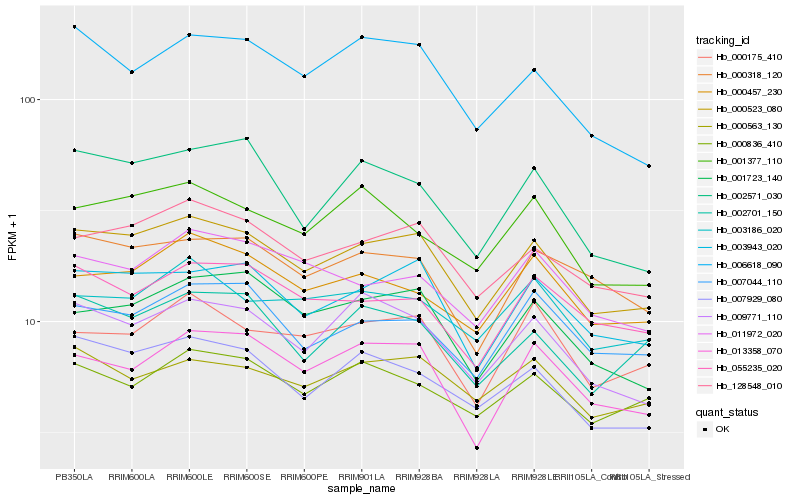
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000523_080 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: beta-catenin-like protein 1 [Jatropha curcas] |
| 2 |
Hb_128548_010 |
0.0456639289 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_003943_020 |
0.0460042887 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase PASTICCINO1 isoform X1 [Jatropha curcas] |
| 4 |
Hb_007929_080 |
0.0578480331 |
- |
- |
PREDICTED: transforming growth factor-beta receptor-associated protein 1 [Jatropha curcas] |
| 5 |
Hb_009771_110 |
0.0614907625 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 2 [Jatropha curcas] |
| 6 |
Hb_001723_140 |
0.0636529469 |
- |
- |
PREDICTED: importin beta-like SAD2 homolog isoform X2 [Jatropha curcas] |
| 7 |
Hb_013358_070 |
0.0653833793 |
- |
- |
protein with unknown function [Ricinus communis] |
| 8 |
Hb_007044_110 |
0.0675112801 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 9 |
Hb_000318_120 |
0.0683358434 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: DNA ligase 1 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000175_410 |
0.0689979124 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 56-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_000457_230 |
0.0693068345 |
- |
- |
PREDICTED: ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplastic [Jatropha curcas] |
| 12 |
Hb_006618_090 |
0.069395013 |
- |
- |
PREDICTED: FAM10 family protein At4g22670 [Jatropha curcas] |
| 13 |
Hb_000563_130 |
0.07006213 |
- |
- |
PREDICTED: WD repeat-containing protein 43 [Jatropha curcas] |
| 14 |
Hb_055235_020 |
0.0705624529 |
- |
- |
PREDICTED: probable mitochondrial saccharopine dehydrogenase-like oxidoreductase At5g39410 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002571_030 |
0.0705882766 |
- |
- |
PREDICTED: microfibrillar-associated protein 1-like [Prunus mume] |
| 16 |
Hb_001377_110 |
0.0707867595 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 31-like [Jatropha curcas] |
| 17 |
Hb_003186_020 |
0.0713255084 |
- |
- |
PREDICTED: histone deacetylase 15 isoform X1 [Jatropha curcas] |
| 18 |
Hb_002701_150 |
0.0726663463 |
- |
- |
PREDICTED: probable tRNA (guanine(26)-N(2))-dimethyltransferase 2 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000836_410 |
0.0727099417 |
- |
- |
sec10, putative [Ricinus communis] |
| 20 |
Hb_011972_020 |
0.0731745809 |
- |
- |
protein binding protein, putative [Ricinus communis] |