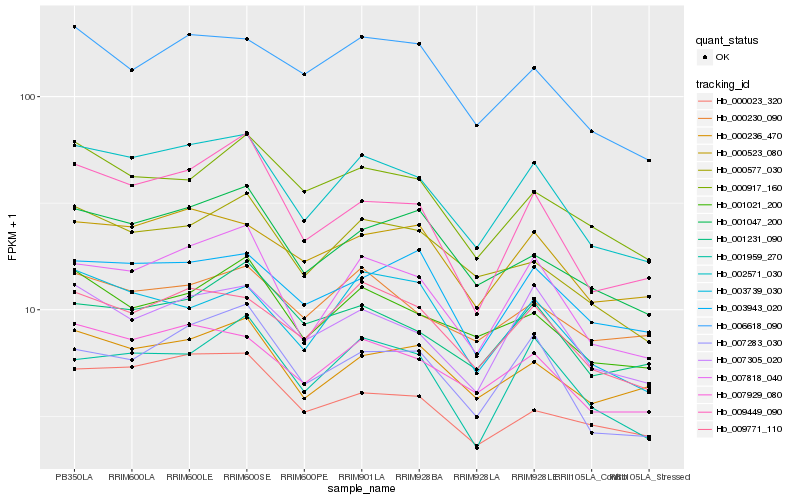
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002571_030 |
0.0 |
- |
- |
PREDICTED: microfibrillar-associated protein 1-like [Prunus mume] |
| 2 |
Hb_007818_040 |
0.0503739434 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 50 isoform X2 [Jatropha curcas] |
| 3 |
Hb_007929_080 |
0.0523524771 |
- |
- |
PREDICTED: transforming growth factor-beta receptor-associated protein 1 [Jatropha curcas] |
| 4 |
Hb_009771_110 |
0.0609865984 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 2 [Jatropha curcas] |
| 5 |
Hb_007305_020 |
0.0658225971 |
- |
- |
PREDICTED: protein SPA1-RELATED 2 [Jatropha curcas] |
| 6 |
Hb_000523_080 |
0.0705882766 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: beta-catenin-like protein 1 [Jatropha curcas] |
| 7 |
Hb_001021_200 |
0.0733033774 |
desease resistance |
Gene Name: AAA |
PREDICTED: peroxisome biogenesis protein 6 [Jatropha curcas] |
| 8 |
Hb_000577_030 |
0.0743504919 |
- |
- |
Ubiquitin carboxyl-terminal hydrolase 12 -like protein [Gossypium arboreum] |
| 9 |
Hb_001047_200 |
0.0746424864 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 18 [Eucalyptus grandis] |
| 10 |
Hb_007283_030 |
0.0748215707 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40C-like [Malus domestica] |
| 11 |
Hb_009449_090 |
0.075164508 |
- |
- |
PREDICTED: RNA-binding protein 25 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000236_470 |
0.0779241343 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g55840 [Jatropha curcas] |
| 13 |
Hb_006618_090 |
0.0780556387 |
- |
- |
PREDICTED: FAM10 family protein At4g22670 [Jatropha curcas] |
| 14 |
Hb_001231_090 |
0.0787790701 |
- |
- |
PREDICTED: CCR4-NOT transcription complex subunit 10 isoform X1 [Jatropha curcas] |
| 15 |
Hb_003739_030 |
0.0813875647 |
- |
- |
PREDICTED: putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX16 [Jatropha curcas] |
| 16 |
Hb_000023_320 |
0.0815080303 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001959_270 |
0.081555942 |
- |
- |
PREDICTED: DNA mismatch repair protein MSH1, mitochondrial isoform X1 [Jatropha curcas] |
| 18 |
Hb_000917_160 |
0.0821579132 |
- |
- |
PREDICTED: protein decapping 5 isoform X1 [Jatropha curcas] |
| 19 |
Hb_003943_020 |
0.0827318737 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase PASTICCINO1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000230_090 |
0.0834933662 |
- |
- |
PREDICTED: WD repeat-containing protein 48 isoform X1 [Jatropha curcas] |