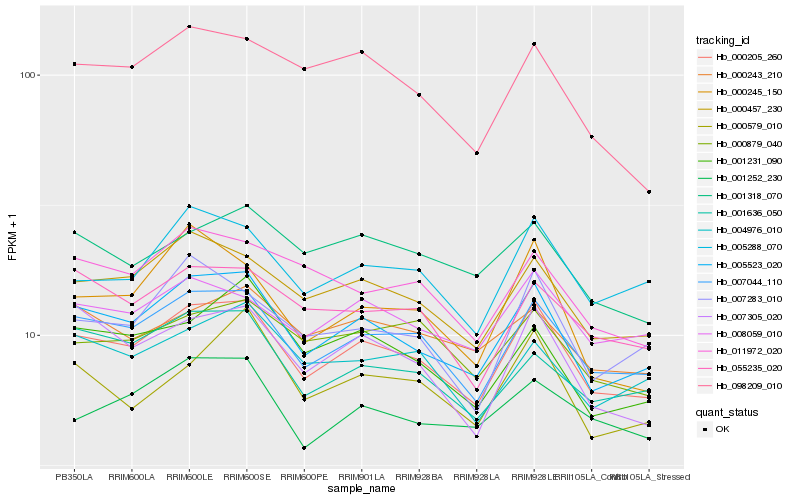
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000205_260 |
0.0 |
transcription factor |
TF Family: SNF2 |
hypothetical protein JCGZ_23567 [Jatropha curcas] |
| 2 |
Hb_005523_020 |
0.0327607593 |
- |
- |
MORPHEUS MOLECULE family protein [Populus trichocarpa] |
| 3 |
Hb_007044_110 |
0.0328647404 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 4 |
Hb_000457_230 |
0.0493645411 |
- |
- |
PREDICTED: ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplastic [Jatropha curcas] |
| 5 |
Hb_005288_070 |
0.0632257068 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 6 |
Hb_007305_020 |
0.065512025 |
- |
- |
PREDICTED: protein SPA1-RELATED 2 [Jatropha curcas] |
| 7 |
Hb_008059_010 |
0.0659984756 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g04810, chloroplastic [Jatropha curcas] |
| 8 |
Hb_055235_020 |
0.068885491 |
- |
- |
PREDICTED: probable mitochondrial saccharopine dehydrogenase-like oxidoreductase At5g39410 isoform X1 [Jatropha curcas] |
| 9 |
Hb_007283_010 |
0.070994072 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40C isoform X1 [Jatropha curcas] |
| 10 |
Hb_000879_040 |
0.0712440309 |
- |
- |
PREDICTED: calcium-transporting ATPase 3, endoplasmic reticulum-type [Jatropha curcas] |
| 11 |
Hb_001636_050 |
0.071669126 |
transcription factor |
TF Family: NF-X1 |
nuclear transcription factor, X-box binding, putative [Ricinus communis] |
| 12 |
Hb_011972_020 |
0.0719146333 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 13 |
Hb_004976_010 |
0.0731920309 |
- |
- |
hypothetical protein RCOM_1429110 [Ricinus communis] |
| 14 |
Hb_001231_090 |
0.0737667291 |
- |
- |
PREDICTED: CCR4-NOT transcription complex subunit 10 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001252_230 |
0.0750829145 |
- |
- |
PREDICTED: host cell factor 2 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001318_070 |
0.0751519974 |
transcription factor |
TF Family: SBP |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000579_010 |
0.0752475883 |
transcription factor |
TF Family: SET |
PREDICTED: probable histone-lysine N-methyltransferase ATXR3 [Jatropha curcas] |
| 18 |
Hb_000243_210 |
0.0755156291 |
- |
- |
protein with unknown function [Ricinus communis] |
| 19 |
Hb_098209_010 |
0.0757689547 |
- |
- |
ATP synthase subunit beta vacuolar, putative [Ricinus communis] |
| 20 |
Hb_000245_150 |
0.0762292704 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |