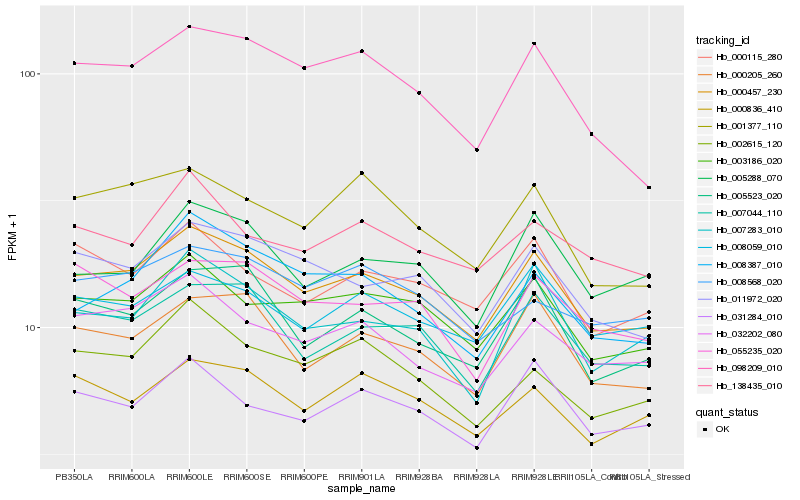
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000457_230 |
0.0 |
- |
- |
PREDICTED: ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplastic [Jatropha curcas] |
| 2 |
Hb_007044_110 |
0.0489446377 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 3 |
Hb_000205_260 |
0.0493645411 |
transcription factor |
TF Family: SNF2 |
hypothetical protein JCGZ_23567 [Jatropha curcas] |
| 4 |
Hb_003186_020 |
0.0517199038 |
- |
- |
PREDICTED: histone deacetylase 15 isoform X1 [Jatropha curcas] |
| 5 |
Hb_011972_020 |
0.0520199651 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 6 |
Hb_005523_020 |
0.0539343302 |
- |
- |
MORPHEUS MOLECULE family protein [Populus trichocarpa] |
| 7 |
Hb_032202_080 |
0.0554879255 |
- |
- |
PREDICTED: peroxisome biogenesis protein 16-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_098209_010 |
0.0613962935 |
- |
- |
ATP synthase subunit beta vacuolar, putative [Ricinus communis] |
| 9 |
Hb_000836_410 |
0.0614129881 |
- |
- |
sec10, putative [Ricinus communis] |
| 10 |
Hb_008059_010 |
0.061569753 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g04810, chloroplastic [Jatropha curcas] |
| 11 |
Hb_007283_010 |
0.0618007076 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40C isoform X1 [Jatropha curcas] |
| 12 |
Hb_055235_020 |
0.0634114367 |
- |
- |
PREDICTED: probable mitochondrial saccharopine dehydrogenase-like oxidoreductase At5g39410 isoform X1 [Jatropha curcas] |
| 13 |
Hb_031284_010 |
0.0638528214 |
- |
- |
PREDICTED: uncharacterized protein LOC105633543 [Jatropha curcas] |
| 14 |
Hb_002615_120 |
0.0640190869 |
- |
- |
PREDICTED: rho GTPase-activating protein 7 isoform X1 [Jatropha curcas] |
| 15 |
Hb_005288_070 |
0.0641976875 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 16 |
Hb_000115_280 |
0.0643121593 |
- |
- |
PREDICTED: glutamine--tRNA ligase [Jatropha curcas] |
| 17 |
Hb_001377_110 |
0.0647625157 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 31-like [Jatropha curcas] |
| 18 |
Hb_008387_010 |
0.0659501797 |
- |
- |
SMAD/FHA domain-containing protein isoform 3 [Theobroma cacao] |
| 19 |
Hb_008568_020 |
0.0674750474 |
- |
- |
PREDICTED: uncharacterized protein LOC105122325 isoform X3 [Populus euphratica] |
| 20 |
Hb_138435_010 |
0.067517258 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
brg-1 associated factor, putative [Ricinus communis] |