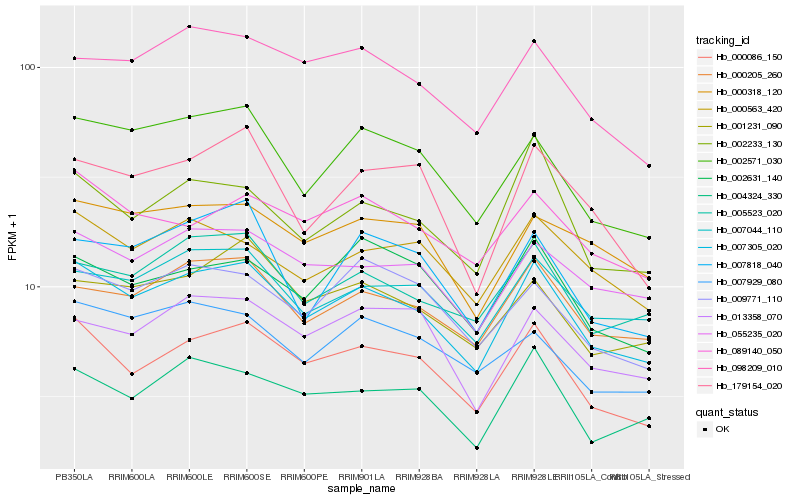
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007305_020 |
0.0 |
- |
- |
PREDICTED: protein SPA1-RELATED 2 [Jatropha curcas] |
| 2 |
Hb_000086_150 |
0.0492965998 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000205_260 |
0.065512025 |
transcription factor |
TF Family: SNF2 |
hypothetical protein JCGZ_23567 [Jatropha curcas] |
| 4 |
Hb_002571_030 |
0.0658225971 |
- |
- |
PREDICTED: microfibrillar-associated protein 1-like [Prunus mume] |
| 5 |
Hb_055235_020 |
0.0714818577 |
- |
- |
PREDICTED: probable mitochondrial saccharopine dehydrogenase-like oxidoreductase At5g39410 isoform X1 [Jatropha curcas] |
| 6 |
Hb_007044_110 |
0.0720017217 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 7 |
Hb_005523_020 |
0.0720073341 |
- |
- |
MORPHEUS MOLECULE family protein [Populus trichocarpa] |
| 8 |
Hb_098209_010 |
0.0761282687 |
- |
- |
ATP synthase subunit beta vacuolar, putative [Ricinus communis] |
| 9 |
Hb_004324_330 |
0.0771104028 |
- |
- |
PREDICTED: uncharacterized protein LOC105648374 [Jatropha curcas] |
| 10 |
Hb_002233_130 |
0.077962797 |
- |
- |
transporter, putative [Ricinus communis] |
| 11 |
Hb_001231_090 |
0.0785549889 |
- |
- |
PREDICTED: CCR4-NOT transcription complex subunit 10 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000563_420 |
0.0799288881 |
- |
- |
PREDICTED: acyl-CoA dehydrogenase family member 10 [Jatropha curcas] |
| 13 |
Hb_000318_120 |
0.0810214911 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: DNA ligase 1 isoform X2 [Jatropha curcas] |
| 14 |
Hb_089140_050 |
0.0813484188 |
- |
- |
hypothetical protein JCGZ_07485 [Jatropha curcas] |
| 15 |
Hb_009771_110 |
0.0813980785 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 2 [Jatropha curcas] |
| 16 |
Hb_002631_140 |
0.0831507191 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 12, chloroplastic [Jatropha curcas] |
| 17 |
Hb_013358_070 |
0.0838813831 |
- |
- |
protein with unknown function [Ricinus communis] |
| 18 |
Hb_007929_080 |
0.0840252805 |
- |
- |
PREDICTED: transforming growth factor-beta receptor-associated protein 1 [Jatropha curcas] |
| 19 |
Hb_179154_020 |
0.0840688107 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 20 |
Hb_007818_040 |
0.0851438713 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 50 isoform X2 [Jatropha curcas] |