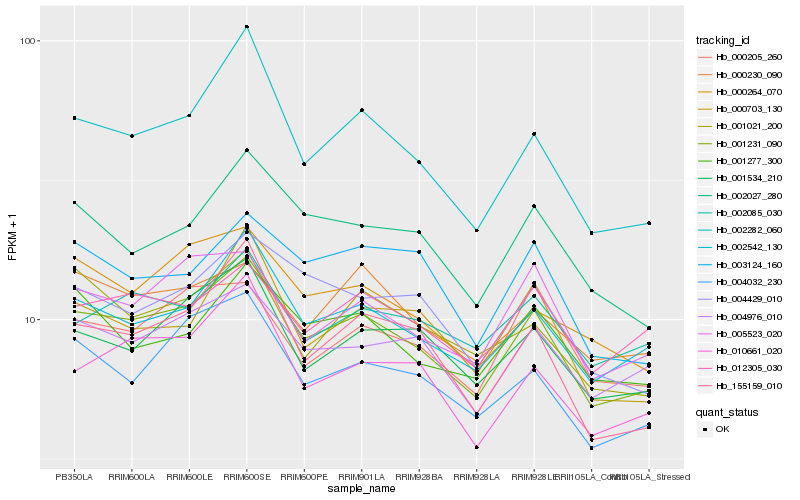
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001231_090 |
0.0 |
- |
- |
PREDICTED: CCR4-NOT transcription complex subunit 10 isoform X1 [Jatropha curcas] |
| 2 |
Hb_002542_130 |
0.0532512471 |
transcription factor |
TF Family: HB |
Homeobox protein LUMINIDEPENDENS, putative [Ricinus communis] |
| 3 |
Hb_002282_060 |
0.0622755027 |
transcription factor |
TF Family: bZIP |
DNA binding protein, putative [Ricinus communis] |
| 4 |
Hb_004976_010 |
0.0624790327 |
- |
- |
hypothetical protein RCOM_1429110 [Ricinus communis] |
| 5 |
Hb_001534_210 |
0.0640424848 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105642316 isoform X2 [Jatropha curcas] |
| 6 |
Hb_004032_230 |
0.0642388698 |
- |
- |
PREDICTED: uncharacterized protein LOC105635143 [Jatropha curcas] |
| 7 |
Hb_005523_020 |
0.0665737192 |
- |
- |
MORPHEUS MOLECULE family protein [Populus trichocarpa] |
| 8 |
Hb_002027_280 |
0.0671144676 |
- |
- |
PREDICTED: sister-chromatid cohesion protein 3 [Jatropha curcas] |
| 9 |
Hb_155159_010 |
0.0676626796 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 1 [Jatropha curcas] |
| 10 |
Hb_004429_010 |
0.0688571618 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 11 |
Hb_010661_020 |
0.0689240972 |
- |
- |
PREDICTED: uncharacterized protein LOC105630767 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000264_070 |
0.0699758206 |
- |
- |
PREDICTED: uncharacterized protein LOC105649513 [Jatropha curcas] |
| 13 |
Hb_001277_300 |
0.0706442617 |
- |
- |
PREDICTED: uncharacterized protein LOC105632051 isoform X4 [Jatropha curcas] |
| 14 |
Hb_012305_030 |
0.0707899287 |
- |
- |
hypothetical protein JCGZ_18979 [Jatropha curcas] |
| 15 |
Hb_000205_260 |
0.0737667291 |
transcription factor |
TF Family: SNF2 |
hypothetical protein JCGZ_23567 [Jatropha curcas] |
| 16 |
Hb_000703_130 |
0.0738766171 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 [Jatropha curcas] |
| 17 |
Hb_003124_160 |
0.0742451227 |
- |
- |
dynamin, putative [Ricinus communis] |
| 18 |
Hb_000230_090 |
0.0744572779 |
- |
- |
PREDICTED: WD repeat-containing protein 48 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002085_030 |
0.0747399246 |
- |
- |
PREDICTED: protein CHROMATIN REMODELING 4 isoform X2 [Jatropha curcas] |
| 20 |
Hb_001021_200 |
0.0754685696 |
desease resistance |
Gene Name: AAA |
PREDICTED: peroxisome biogenesis protein 6 [Jatropha curcas] |