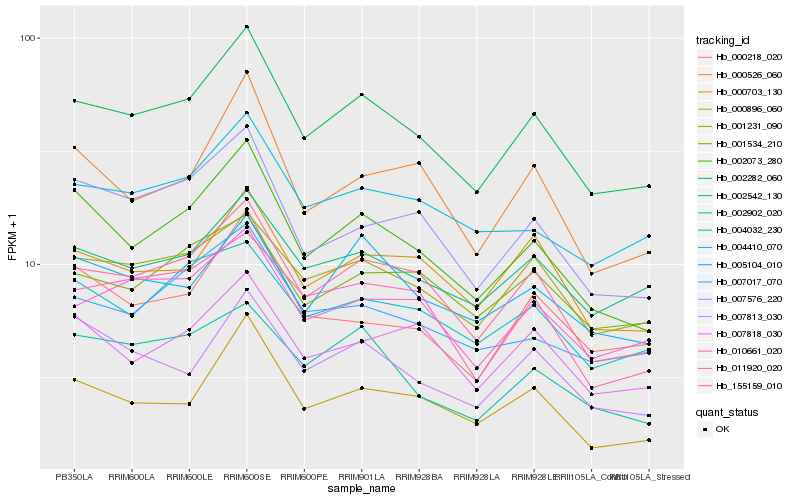
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002282_060 |
0.0 |
transcription factor |
TF Family: bZIP |
DNA binding protein, putative [Ricinus communis] |
| 2 |
Hb_155159_010 |
0.0445369093 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 1 [Jatropha curcas] |
| 3 |
Hb_001231_090 |
0.0622755027 |
- |
- |
PREDICTED: CCR4-NOT transcription complex subunit 10 isoform X1 [Jatropha curcas] |
| 4 |
Hb_002073_280 |
0.063624024 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 3 isoform X1 [Jatropha curcas] |
| 5 |
Hb_010661_020 |
0.0675198084 |
- |
- |
PREDICTED: uncharacterized protein LOC105630767 isoform X1 [Jatropha curcas] |
| 6 |
Hb_002542_130 |
0.067638109 |
transcription factor |
TF Family: HB |
Homeobox protein LUMINIDEPENDENS, putative [Ricinus communis] |
| 7 |
Hb_000703_130 |
0.0739351542 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 [Jatropha curcas] |
| 8 |
Hb_004410_070 |
0.0746376165 |
- |
- |
PREDICTED: protein LTV1 homolog [Jatropha curcas] |
| 9 |
Hb_000896_060 |
0.0772401227 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 10 |
Hb_007576_220 |
0.0780158437 |
- |
- |
PREDICTED: kanadaptin [Jatropha curcas] |
| 11 |
Hb_005104_010 |
0.0784844557 |
- |
- |
PREDICTED: uncharacterized protein LOC105140796 isoform X1 [Populus euphratica] |
| 12 |
Hb_004032_230 |
0.0791643481 |
- |
- |
PREDICTED: uncharacterized protein LOC105635143 [Jatropha curcas] |
| 13 |
Hb_007017_070 |
0.0796907591 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 3 isoform X1 [Jatropha curcas] |
| 14 |
Hb_011920_020 |
0.0799122647 |
- |
- |
PREDICTED: uncharacterized protein LOC105645397 [Jatropha curcas] |
| 15 |
Hb_001534_210 |
0.0807026931 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105642316 isoform X2 [Jatropha curcas] |
| 16 |
Hb_007818_030 |
0.0814437904 |
- |
- |
PREDICTED: transcriptional corepressor SEUSS-like [Jatropha curcas] |
| 17 |
Hb_007813_030 |
0.0826151868 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At4g19050 [Jatropha curcas] |
| 18 |
Hb_002902_020 |
0.0837761351 |
- |
- |
PREDICTED: uncharacterized protein LOC105639827 [Jatropha curcas] |
| 19 |
Hb_000218_020 |
0.084094533 |
- |
- |
synaptosomal associated protein, putative [Ricinus communis] |
| 20 |
Hb_000526_060 |
0.0841007244 |
- |
- |
PREDICTED: clustered mitochondria protein [Jatropha curcas] |