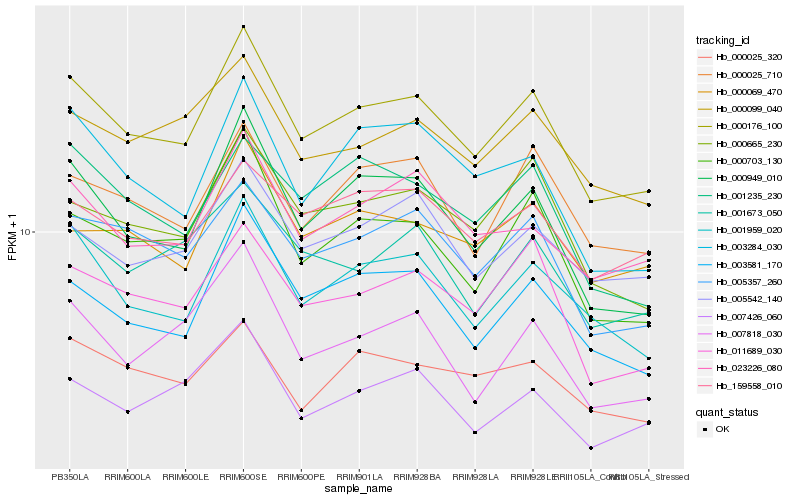
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000176_100 |
0.0 |
- |
- |
PREDICTED: probable splicing factor 3A subunit 1 [Solanum lycopersicum] |
| 2 |
Hb_000949_010 |
0.0520468248 |
- |
- |
PREDICTED: regulator of nonsense transcripts UPF2 [Jatropha curcas] |
| 3 |
Hb_001959_020 |
0.0523902819 |
- |
- |
PREDICTED: uncharacterized protein LOC105638454 isoform X1 [Jatropha curcas] |
| 4 |
Hb_003581_170 |
0.054004024 |
- |
- |
PREDICTED: alpha-amylase 3, chloroplastic [Jatropha curcas] |
| 5 |
Hb_005357_260 |
0.0542458753 |
- |
- |
PREDICTED: polyadenylation and cleavage factor homolog 4 [Jatropha curcas] |
| 6 |
Hb_000703_130 |
0.0550186169 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 [Jatropha curcas] |
| 7 |
Hb_011689_030 |
0.056682363 |
- |
- |
80 kD MCM3-associated protein, putative [Ricinus communis] |
| 8 |
Hb_000665_230 |
0.058653647 |
- |
- |
PREDICTED: uncharacterized protein LOC105637615 [Jatropha curcas] |
| 9 |
Hb_023226_080 |
0.0595637915 |
transcription factor |
TF Family: C3H |
PREDICTED: 30-kDa cleavage and polyadenylation specificity factor 30 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000025_710 |
0.0618062793 |
transcription factor |
TF Family: SNF2 |
Chromodomain-helicase-DNA-binding 2 [Gossypium arboreum] |
| 11 |
Hb_005542_140 |
0.0624354369 |
- |
- |
PREDICTED: U-box domain-containing protein 62-like [Jatropha curcas] |
| 12 |
Hb_000099_040 |
0.0630421809 |
- |
- |
unnamed protein product [Coffea canephora] |
| 13 |
Hb_001673_050 |
0.0651319599 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 8-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_007426_060 |
0.0656613291 |
desease resistance |
Gene Name: NB-ARC |
unnamed protein product [Coffea canephora] |
| 15 |
Hb_000069_470 |
0.0671741019 |
- |
- |
PREDICTED: uncharacterized protein LOC105642863 isoform X1 [Jatropha curcas] |
| 16 |
Hb_003284_030 |
0.0674665828 |
transcription factor |
TF Family: Orphans |
PREDICTED: paired amphipathic helix protein Sin3-like 2 isoform X1 [Jatropha curcas] |
| 17 |
Hb_159558_010 |
0.0675159573 |
- |
- |
PREDICTED: uncharacterized protein LOC105635846 [Jatropha curcas] |
| 18 |
Hb_001235_230 |
0.0689442612 |
- |
- |
PREDICTED: protein transport protein SEC16B homolog [Jatropha curcas] |
| 19 |
Hb_000025_320 |
0.0692595156 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g28010 [Jatropha curcas] |
| 20 |
Hb_007818_030 |
0.0693113278 |
- |
- |
PREDICTED: transcriptional corepressor SEUSS-like [Jatropha curcas] |