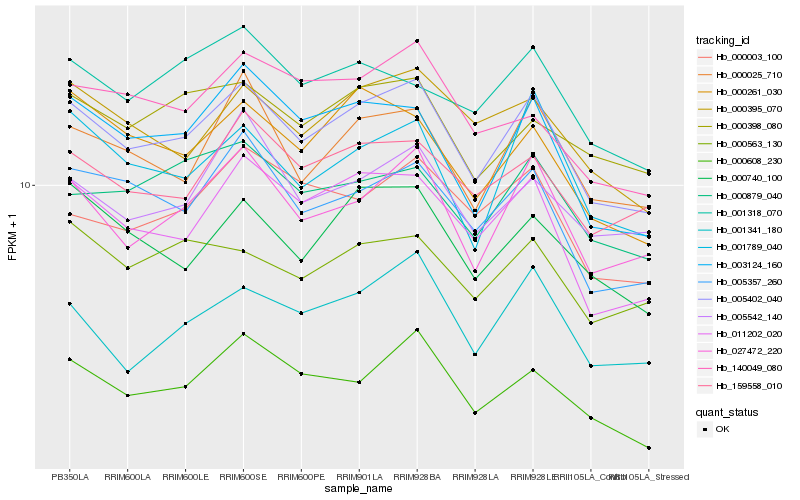
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005402_040 |
0.0 |
- |
- |
PREDICTED: replication factor C subunit 1 [Jatropha curcas] |
| 2 |
Hb_027472_220 |
0.0511046361 |
- |
- |
PREDICTED: nucleolar complex protein 3 homolog isoform X1 [Jatropha curcas] |
| 3 |
Hb_011202_020 |
0.0534439587 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 3 isoform X1 [Jatropha curcas] |
| 4 |
Hb_005357_260 |
0.0582043201 |
- |
- |
PREDICTED: polyadenylation and cleavage factor homolog 4 [Jatropha curcas] |
| 5 |
Hb_003124_160 |
0.0590417149 |
- |
- |
dynamin, putative [Ricinus communis] |
| 6 |
Hb_000025_710 |
0.0601328658 |
transcription factor |
TF Family: SNF2 |
Chromodomain-helicase-DNA-binding 2 [Gossypium arboreum] |
| 7 |
Hb_000395_070 |
0.0626589354 |
- |
- |
PREDICTED: serine/threonine-protein kinase BLUS1 isoform X2 [Jatropha curcas] |
| 8 |
Hb_159558_010 |
0.0628283636 |
- |
- |
PREDICTED: uncharacterized protein LOC105635846 [Jatropha curcas] |
| 9 |
Hb_000740_100 |
0.0640833185 |
- |
- |
calpain, putative [Ricinus communis] |
| 10 |
Hb_000261_030 |
0.0649101666 |
- |
- |
PREDICTED: uncharacterized protein LOC105633147 [Jatropha curcas] |
| 11 |
Hb_000608_230 |
0.0661016154 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Jatropha curcas] |
| 12 |
Hb_001789_040 |
0.0677046925 |
- |
- |
PREDICTED: uncharacterized protein LOC105648457 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000879_040 |
0.0678723929 |
- |
- |
PREDICTED: calcium-transporting ATPase 3, endoplasmic reticulum-type [Jatropha curcas] |
| 14 |
Hb_000398_080 |
0.0679172693 |
- |
- |
tip120, putative [Ricinus communis] |
| 15 |
Hb_001341_180 |
0.0679856406 |
- |
- |
PREDICTED: transmembrane protein 209 [Jatropha curcas] |
| 16 |
Hb_005542_140 |
0.0686742468 |
- |
- |
PREDICTED: U-box domain-containing protein 62-like [Jatropha curcas] |
| 17 |
Hb_000003_100 |
0.0689199971 |
transcription factor |
TF Family: SNF2 |
PREDICTED: transcription termination factor 2 isoform X5 [Jatropha curcas] |
| 18 |
Hb_000563_130 |
0.0697461837 |
- |
- |
PREDICTED: WD repeat-containing protein 43 [Jatropha curcas] |
| 19 |
Hb_001318_070 |
0.0709918582 |
transcription factor |
TF Family: SBP |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_140049_080 |
0.0710243104 |
- |
- |
PREDICTED: probable RNA helicase SDE3 [Jatropha curcas] |