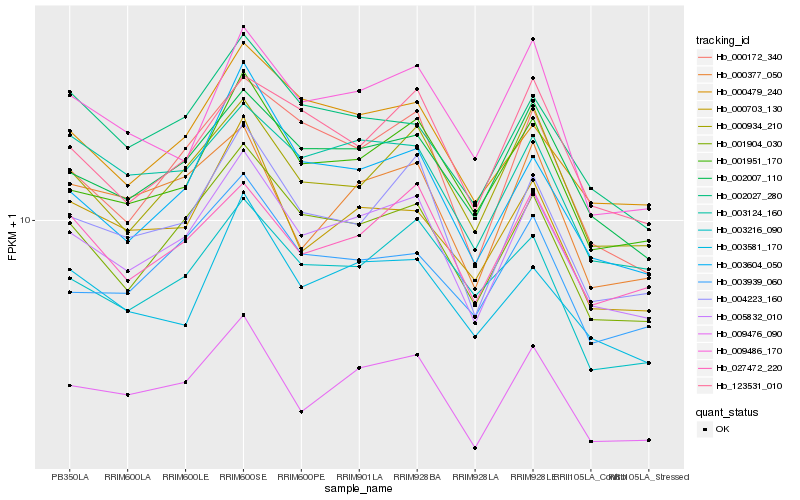
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005832_010 |
0.0 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001904_030 |
0.0440085047 |
- |
- |
PREDICTED: importin-5 [Jatropha curcas] |
| 3 |
Hb_004223_160 |
0.0470055167 |
- |
- |
PREDICTED: 5'-3' exoribonuclease 3 isoform X2 [Jatropha curcas] |
| 4 |
Hb_009476_090 |
0.0473056435 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 1 [Jatropha curcas] |
| 5 |
Hb_002007_110 |
0.0531701508 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ25 isoform X2 [Jatropha curcas] |
| 6 |
Hb_001951_170 |
0.0545175932 |
- |
- |
PREDICTED: uncharacterized protein LOC105649755 [Jatropha curcas] |
| 7 |
Hb_000172_340 |
0.0565852473 |
- |
- |
PREDICTED: uncharacterized protein LOC101261327 [Solanum lycopersicum] |
| 8 |
Hb_000934_210 |
0.0616218268 |
- |
- |
suppressor of ty, putative [Ricinus communis] |
| 9 |
Hb_003604_050 |
0.0634570001 |
- |
- |
PREDICTED: MAP3K epsilon protein kinase 1-like isoform X2 [Jatropha curcas] |
| 10 |
Hb_000479_240 |
0.0635702892 |
- |
- |
PREDICTED: probable ubiquitin conjugation factor E4 isoform X2 [Jatropha curcas] |
| 11 |
Hb_003124_160 |
0.064368675 |
- |
- |
dynamin, putative [Ricinus communis] |
| 12 |
Hb_003581_170 |
0.0660471979 |
- |
- |
PREDICTED: alpha-amylase 3, chloroplastic [Jatropha curcas] |
| 13 |
Hb_027472_220 |
0.0668326505 |
- |
- |
PREDICTED: nucleolar complex protein 3 homolog isoform X1 [Jatropha curcas] |
| 14 |
Hb_002027_280 |
0.0683057367 |
- |
- |
PREDICTED: sister-chromatid cohesion protein 3 [Jatropha curcas] |
| 15 |
Hb_123531_010 |
0.0684666255 |
- |
- |
PREDICTED: nuclear pore complex protein NUP98A isoform X1 [Jatropha curcas] |
| 16 |
Hb_000703_130 |
0.0694425286 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 [Jatropha curcas] |
| 17 |
Hb_003939_060 |
0.070363939 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_003216_090 |
0.0712774511 |
- |
- |
PREDICTED: nuclear pore complex protein NUP133 isoform X1 [Jatropha curcas] |
| 19 |
Hb_009486_170 |
0.0721482391 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 4G [Jatropha curcas] |
| 20 |
Hb_000377_050 |
0.0723771939 |
- |
- |
PREDICTED: sister chromatid cohesion protein PDS5 homolog A isoform X1 [Jatropha curcas] |