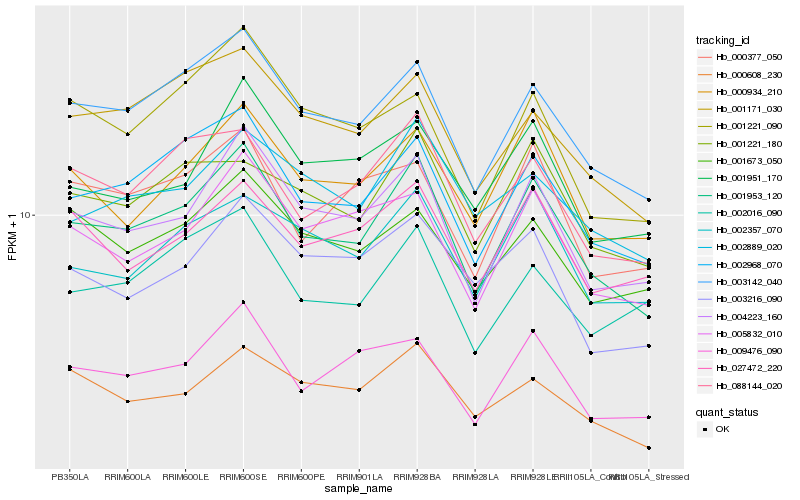
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001953_120 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105648761 isoform X1 [Jatropha curcas] |
| 2 |
Hb_003142_040 |
0.0539545312 |
- |
- |
Sf3a3 [Gossypium arboreum] |
| 3 |
Hb_002968_070 |
0.055244609 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 4b isoform X2 [Jatropha curcas] |
| 4 |
Hb_004223_160 |
0.0556900935 |
- |
- |
PREDICTED: 5'-3' exoribonuclease 3 isoform X2 [Jatropha curcas] |
| 5 |
Hb_088144_020 |
0.0726497478 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 6-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_005832_010 |
0.0753040157 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000934_210 |
0.0760626778 |
- |
- |
suppressor of ty, putative [Ricinus communis] |
| 8 |
Hb_009476_090 |
0.076138265 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 1 [Jatropha curcas] |
| 9 |
Hb_001171_030 |
0.0762325854 |
transcription factor |
TF Family: LUG |
PREDICTED: transcriptional corepressor LEUNIG-like [Jatropha curcas] |
| 10 |
Hb_001221_180 |
0.0777095268 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 11 |
Hb_000608_230 |
0.0797304687 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Jatropha curcas] |
| 12 |
Hb_002357_070 |
0.0803274199 |
transcription factor |
TF Family: PHD |
PREDICTED: protein strawberry notch homolog 1 [Jatropha curcas] |
| 13 |
Hb_000377_050 |
0.0830130831 |
- |
- |
PREDICTED: sister chromatid cohesion protein PDS5 homolog A isoform X1 [Jatropha curcas] |
| 14 |
Hb_001951_170 |
0.0852744562 |
- |
- |
PREDICTED: uncharacterized protein LOC105649755 [Jatropha curcas] |
| 15 |
Hb_027472_220 |
0.0858736069 |
- |
- |
PREDICTED: nucleolar complex protein 3 homolog isoform X1 [Jatropha curcas] |
| 16 |
Hb_002889_020 |
0.0869901393 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 3 [Jatropha curcas] |
| 17 |
Hb_003216_090 |
0.0881459911 |
- |
- |
PREDICTED: nuclear pore complex protein NUP133 isoform X1 [Jatropha curcas] |
| 18 |
Hb_002016_090 |
0.0882356592 |
- |
- |
PREDICTED: DNA mismatch repair protein PMS1 [Jatropha curcas] |
| 19 |
Hb_001221_090 |
0.0884285105 |
- |
- |
PREDICTED: uncharacterized protein LOC105640211 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001673_050 |
0.0884440132 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 8-like isoform X1 [Jatropha curcas] |