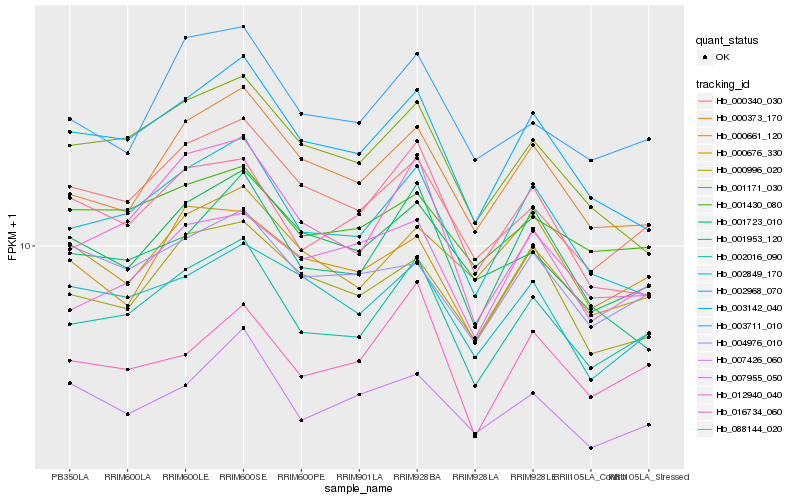
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002016_090 |
0.0 |
- |
- |
PREDICTED: DNA mismatch repair protein PMS1 [Jatropha curcas] |
| 2 |
Hb_002968_070 |
0.057497853 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 4b isoform X2 [Jatropha curcas] |
| 3 |
Hb_000340_030 |
0.0661319529 |
- |
- |
PREDICTED: myosin heavy chain, non-muscle-like [Jatropha curcas] |
| 4 |
Hb_003142_040 |
0.0695520967 |
- |
- |
Sf3a3 [Gossypium arboreum] |
| 5 |
Hb_012940_040 |
0.0705366206 |
- |
- |
Splicing factor, arginine/serine-rich, putative [Ricinus communis] |
| 6 |
Hb_000373_170 |
0.070656826 |
- |
- |
PREDICTED: exocyst complex component SEC5A-like [Jatropha curcas] |
| 7 |
Hb_003711_010 |
0.0729040556 |
- |
- |
isopropylmalate synthase, putative [Ricinus communis] |
| 8 |
Hb_001723_010 |
0.0798441537 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL6 [Jatropha curcas] |
| 9 |
Hb_016734_060 |
0.0810527134 |
- |
- |
PREDICTED: nucleolar protein 6 [Jatropha curcas] |
| 10 |
Hb_000996_020 |
0.0819827543 |
- |
- |
PREDICTED: RNA-binding protein NOB1 [Jatropha curcas] |
| 11 |
Hb_000661_120 |
0.0820139624 |
- |
- |
cap binding protein, putative [Ricinus communis] |
| 12 |
Hb_001171_030 |
0.0822835088 |
transcription factor |
TF Family: LUG |
PREDICTED: transcriptional corepressor LEUNIG-like [Jatropha curcas] |
| 13 |
Hb_001430_080 |
0.0827065897 |
- |
- |
PREDICTED: putative mediator of RNA polymerase II transcription subunit 26 [Jatropha curcas] |
| 14 |
Hb_088144_020 |
0.0836081766 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 6-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_002849_170 |
0.0837178921 |
- |
- |
PREDICTED: uncharacterized protein LOC105642955 isoform X1 [Jatropha curcas] |
| 16 |
Hb_007426_060 |
0.0841640899 |
desease resistance |
Gene Name: NB-ARC |
unnamed protein product [Coffea canephora] |
| 17 |
Hb_007955_050 |
0.0876596256 |
- |
- |
PREDICTED: multiple RNA-binding domain-containing protein 1 [Jatropha curcas] |
| 18 |
Hb_001953_120 |
0.0882356592 |
- |
- |
PREDICTED: uncharacterized protein LOC105648761 isoform X1 [Jatropha curcas] |
| 19 |
Hb_004976_010 |
0.0891450911 |
- |
- |
hypothetical protein RCOM_1429110 [Ricinus communis] |
| 20 |
Hb_000676_330 |
0.0892749212 |
- |
- |
protein with unknown function [Ricinus communis] |