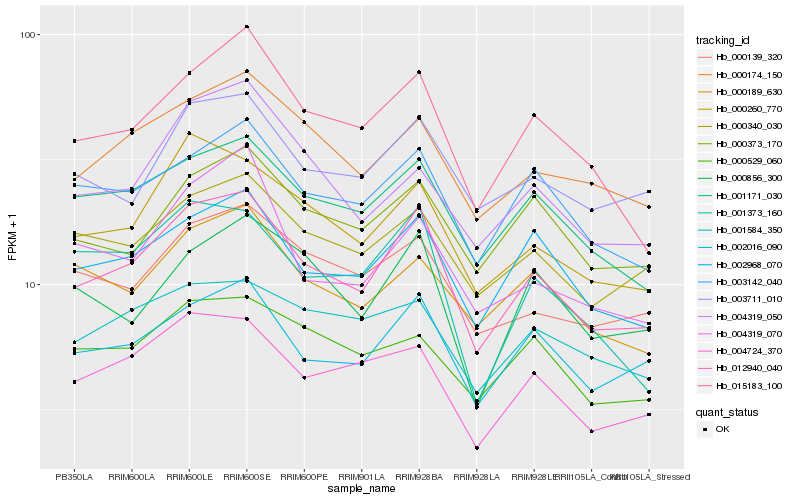
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012940_040 |
0.0 |
- |
- |
Splicing factor, arginine/serine-rich, putative [Ricinus communis] |
| 2 |
Hb_002968_070 |
0.0699176247 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 4b isoform X2 [Jatropha curcas] |
| 3 |
Hb_002016_090 |
0.0705366206 |
- |
- |
PREDICTED: DNA mismatch repair protein PMS1 [Jatropha curcas] |
| 4 |
Hb_000260_770 |
0.071539885 |
- |
- |
er lumen protein retaining receptor, putative [Ricinus communis] |
| 5 |
Hb_001171_030 |
0.0736986195 |
transcription factor |
TF Family: LUG |
PREDICTED: transcriptional corepressor LEUNIG-like [Jatropha curcas] |
| 6 |
Hb_015183_100 |
0.0756090485 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360 [Jatropha curcas] |
| 7 |
Hb_000174_150 |
0.0769497672 |
- |
- |
stearoyl-acyl-carrier protein desaturase [Ricinus communis] |
| 8 |
Hb_004724_370 |
0.0771058705 |
- |
- |
PREDICTED: mRNA-decapping enzyme-like protein [Jatropha curcas] |
| 9 |
Hb_003142_040 |
0.0785041593 |
- |
- |
Sf3a3 [Gossypium arboreum] |
| 10 |
Hb_000340_030 |
0.0833328983 |
- |
- |
PREDICTED: myosin heavy chain, non-muscle-like [Jatropha curcas] |
| 11 |
Hb_000139_320 |
0.0846966082 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF1A [Jatropha curcas] |
| 12 |
Hb_003711_010 |
0.0848956993 |
- |
- |
isopropylmalate synthase, putative [Ricinus communis] |
| 13 |
Hb_000373_170 |
0.0870317881 |
- |
- |
PREDICTED: exocyst complex component SEC5A-like [Jatropha curcas] |
| 14 |
Hb_000189_630 |
0.0894665547 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 15 |
Hb_000856_300 |
0.0924412691 |
- |
- |
PREDICTED: uncharacterized protein LOC105640498 [Jatropha curcas] |
| 16 |
Hb_001584_350 |
0.0932099562 |
- |
- |
hypothetical protein JCGZ_23178 [Jatropha curcas] |
| 17 |
Hb_001373_160 |
0.0938622633 |
- |
- |
hypothetical protein JCGZ_16907 [Jatropha curcas] |
| 18 |
Hb_000529_060 |
0.0944516107 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 17 isoform X1 [Jatropha curcas] |
| 19 |
Hb_004319_050 |
0.0947035969 |
- |
- |
PREDICTED: senescence-associated carboxylesterase 101-like [Populus euphratica] |
| 20 |
Hb_004319_070 |
0.0962437643 |
- |
- |
hypothetical protein RCOM_0852460 [Ricinus communis] |